The intercontinental disjunctions between eastern Asia and North America have attracted much attention from evolutionary biologists in the last two decades. Biogeographic analyses of ferns with an eastern Asian-North American disjunction are few. The Adiantum pedatum complex includes four species distributed in North America and eastern Asia extending to the Himalayas (Fig. 1). The monophyly of the complex needs to be tested and diversification history of the four species needs to be reconstructed.
Recently, Dr. LU Jinmei, Profs. LI Dezhu and WEN Jun analyzed Plastid (atpA, atpB, rbcL, trnL-F, and rps4-trnS) sequences of 100 accessions representing the biogeographic diversity of Adiantum. The results suggested that the A. pedatum complex is monophyletic and sister to the eastern Asian A. edentulum (Fig. 2). Accessions of A. pedatum do not form a clade; instead three subgroups are recognizable. The clade of A. aleuticum and A. viridimontanum is nested within A. pedatum. The Asian A. myriosorum is sister to the A. pedatum-A. aleuticum clade. Both DIVA and LAGRANGE analyses suggest an eastern Asian origin of the A. pedatum complex. The age of the crown A. pedatum complex is dated to be at 4.27 (2.24–6.57) million years ago. The results implied that the currently recognized eastern Asian–North American disjunct species A. pedatum needs to be segregated into three species, corresponding to populations in eastern North America, China, and Japan. The eastern Asian–North American disjunction in the complex is inferred to be the result of two intercontinental migrations, one from eastern Asia into North America in the late Tertiary and the other from North America back to eastern Asia in the Pleistocene.
The paper “Biogeographic disjunction between eastern Asia and North America in the Adiantum pedatum complex (Pteridaceae)” was published on “American Journal of Botany”. http://www.amjbot.org/cgi/content/abstract/98/10/1680.
The study was supported by grants from National Basic Research Program of China (973 Program, 2007CB411601), the National Natural Science Foundation of China (31070199), the Project of Knowledge Innovation Program of the Chinese Academy of Sciences (2010KIBA02), and the Research Fund for the Large-Scale Scientific Facilities of the Chinese Academy of Sciences (2009-LSF-GBOWS-01).
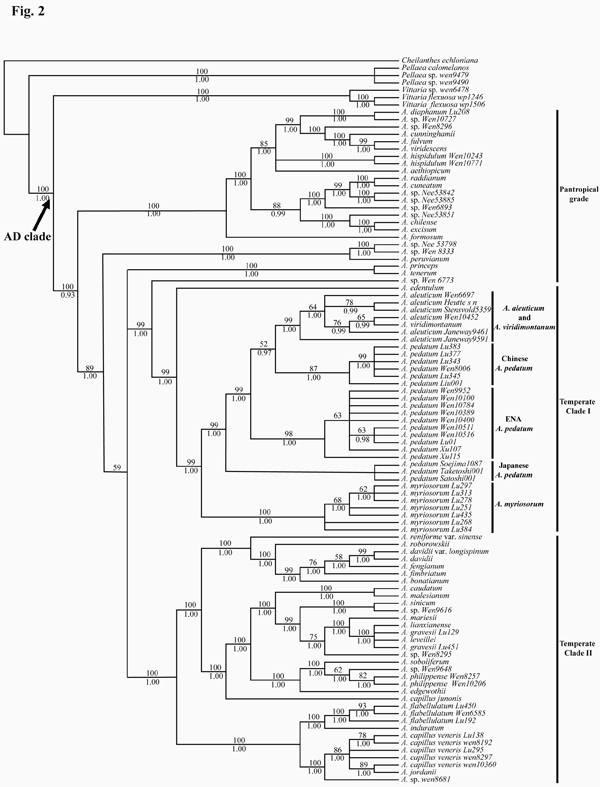
Fig. 2. Strict consensus tree of four maximally parsimonious trees derived from the analysis of atpA , atpB , rbcL , trnL-F , and rps4 - trnS sequences. The bootstrap values for 1000 replicates are shown above the branches, the Bayesian posterior probabilities are shown below the branches. ENA, eastern North America. (image by LU Jinmei)




