Compared to their counterparts in animals, the mitochondrial (mt) genomes of angiosperms exhibit a number of unique features, such as expanded genome size, a generally low rate of molecular evolution, and frequent structure rearrangement via recombination. However, unraveling angiosperm mt genomes evolution is hindered by the few completed genomes, and the established sequencing approaches are time- and labor-intensive. Next-generation sequencing that is not only high-throughput but also low-cost has already revolutionized approaches for genome sequencing, including angiosperm chloroplast genomes. However, it is just beginning to be applied to angiosperm mt genomes.
Recently, researchers of Molecular Phylogenetics and Biogeography Group of the Kunming Institute of Botany of the Chinese Academy of Sciences have completed six woody bamboos chloroplast genomes using Illumina sequencing. Meanwhile, a large number of sequence reads were found in the total reads being of mtDNA origin. Could the mt genome be assembled from these reads at the same time? And if so, this would be a rapid and efficient approach to sequence angiosperm mt genomes. With combination of de novo and reference-guided assembly, 39.5-fold coverage Illumina reads from one of six bamboos, Ferrocalamus rimosivaginus, were finally assembled into 13 scaffolds and one contig totaling 432,839 bp. The assembled genome contains nearly the same genes as the completed mt genomes in other members of the grass family (Poaceae). Additionally, the assembled sequences were validated by conventional Sanger sequencing. The result demonstrated that this was a rapid and efficient approach to obtain angiosperm mt genome sequences.
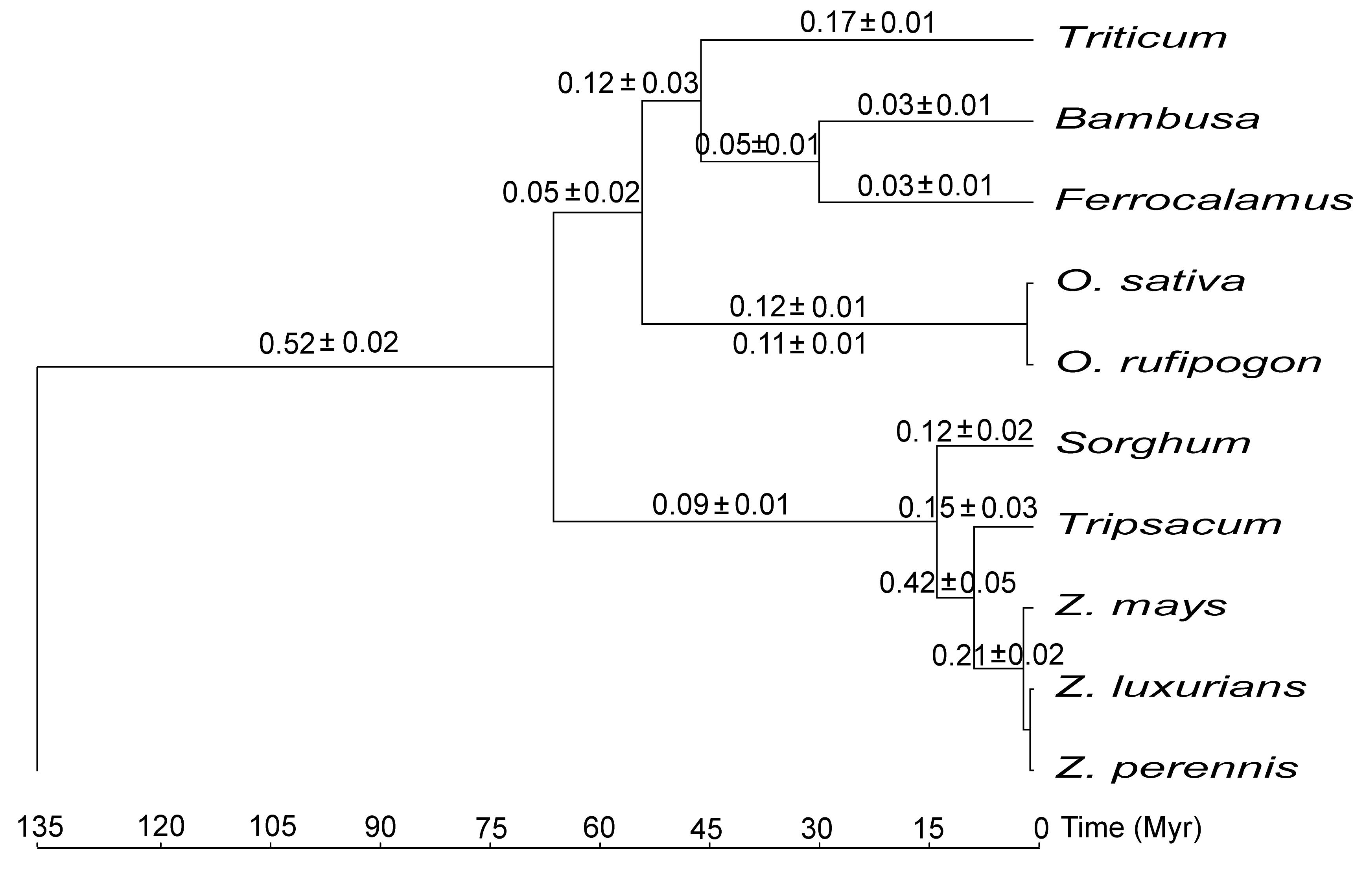
For examining evolutionary rate in grass mt genomes, a phylogenetic tree including 22 taxa based on 31 mt genes was reconstructed. The topology of the well-resolved tree was almost identical to that inferred from chloroplast genome with only minor difference. And the inconsistency possibly derived from long branch attraction (LBA) in mtDNA tree. By calculating absolute substitution rates, significant rate change (~4-fold) in mt genome before and after diversification of Poaceae was detected. Furthermore, the rate change was correlated with that of chloroplast genomes in grasses.