Arundinarieae is a tribe comprising about 600 taxa in 32 genera and belongs to the family Poaceae, subfamily Bambusoideae. Bamboos in this tribe are mainly distributed in the temperate and subtropical regions of Africa, Asia, and southeastern North America, with East Asia as the center of diversity with more than 500 species. Most of these species are important ecological components of the subtropical and temperate forests. A number of species of Fargesia and Yushania are the main food of the giant panda. Some species, such as Moso bamboo (Phyllostachys edulis), have been cultivated for a long history with great economic value in China and adjacent regions. Arundinarieae is a highly diversified group with complex features of morphology, including pachymorph or leptomorph rhizomes, solitary to many branches, semelauctant or iterauctant synflorescences, 3–6 stamens, and bacoid, nucoid or basic caryopsis. This tribe is notorious for being taxonomically extremely difficult.
The research group of Professor LI Dezhu of Kunming Institute of Botany, Chinese Academy of Sciences has been focusing on the molecular phylogeny and evolution of Bambusoideae for many years. Recently, they reconstructed the phylogeny of Arundinarieae using eight non-coding plastid regions for 146 species in 26 genera, and ten major lineages were recovered in this tribe with interrelationships unresolved which were significantly different from the morphological classification (Molecular Phylogenetics and Evolution, 2010. 56: 821-839.). Subsequently, they exploited the Illumina technology to sequence the chloroplast and mitochondrial genomes of several species in this tribe, and reconstructed the phylogenetic trees by phylogenomic methods (PLoS ONE, 2011. 6(5): e20596; PLoS ONE, 2012. 7(1): e30297).
To further illuminate the complex evolutionary history of Arundinarieae, Drs. ZHANG Yuxiao and ZENG Chunxia performed the phylogenetic reconstruction of 108 species in 25 genera of Arundinarieae with the nuclear GBSSI gene. Thirteen major clades were revealed, which had better resolution at the generic level than the plastid results. However, conspicuous conflicts were revealed between the plastid and nuclear trees. The patterns of incongruence suggested that lack of informative characters, incomplete lineage sorting, and/or hybridization (introgression) could be the causes. Seven putative hybrid species were hypothesized, four of which were discussed in detail on the basis of topological incongruence, chromosome numbers, morphology, and distribution patterns. Overall, those studies indicate that the tribe Arundinarieae has undergone a complex evolution.
This study has been recently published in Molecular Phylogenetics and Evolution (2012. 63: 777–797)(http://www.sciencedirect.com/science/article/pii/S1055790312000760). It was funded by the National Natural Science Foundation of China (Grant Nos.: 31100148 & No. 31100165), also partly supported by the Project of Knowledge innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-EW-J-24).
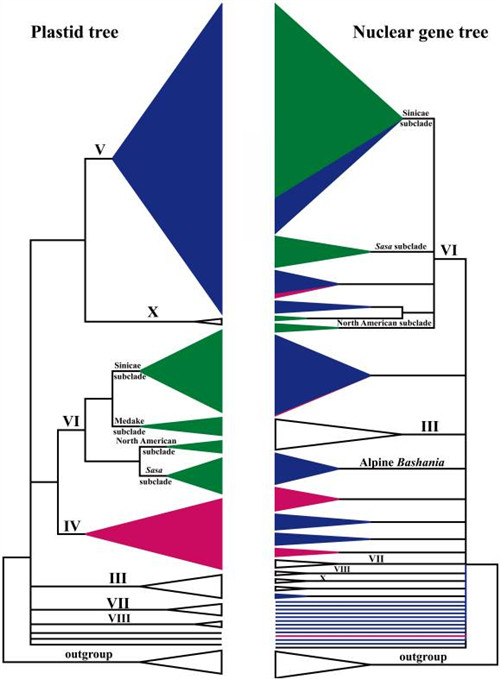
Simplified version of Fig. 1 (left) and Fig. 2 (right), which denotes the major differences between the plastid and nuclear gene trees.
The clade names correspond to those in Fig. 1 and Fig. 2. (Image by KIB)




