In a study that offers clues why tea is so popular worldwide, Chinese researchers announced Monday they have successfully sequenced the genome of the evergreen shrub Camellia sinensis, known as tea tree, for the first time.
The genus Camellia contains over 100 species, but the most popular varieties of tea, including black tea, green tea, Oolong tea, white tea, and chai, all come from the leaves of the evergreen shrub Camellia sinensis.
"There are many diverse flavors, but the mystery is what determines or what is the genetic basis of tea flavors?" GAO Lizhi, plant geneticist of Kunming Institute of Botany in China and senior author of the study published in the journal Molecular Plant, told Xinhua. "We believe that sequencing tea tree genome would help to solve these problems."
GAO, who got training in genomics and bioinformatics in the United States,became the first scientist to do such a job in 2010, and even with modern sequencing, assembling the genome took his team over five years.
It turned out that the tea tree genome is much larger than initially expected. At 3.02 billion base pairs in length, it is more than four times the size of the coffee plant genome and much larger than most sequenced plant species.
GAO estimated that more than half of the tea tree genome are made of "jumping genes", which have copied-and-pasted themselves into different spots in the genome numerous times.
This resulted in a dramatic expansion in genome size of tea tree, which could have helped the plant adapt to different climates and environmental stresses in Asia, Europe, Africa, the Americas and Oceania, he said.
Previous studies have suggested that tea owes much of its flavor to caffeine, an amino acid called theanine, and a group of antioxidants called flavonoids, including a bitter-tasting one called catechin.
GAO's team found that tea tree leaves not only contain high levels of catechins, caffeine and other flavonoids, but also have multiple copies of the genes that produce caffeine and flavonoids.
But they observed no significant differents in theanine content between tea tree and other species in the Camellia family that are unsuitable for making teas.
"This answers why leaves from some well-known camellias with their attractive flowers and the traditional oil tree Camellia oleifera can not be used to make tea, (that's) because of significantly low production of catechins and caffeine but not theanine," Gao said.
"In other words, expression levels of most flavonoid and caffeine but not theanine-related genes determines the tea processing suitability."
Tea is among the world's most oldest and important nonalcoholic caffeine-containing beverage, and tea tree was originally domesticated in Southwest China. It's estimated that some three billion people worldwide drink tea.
"So, our achievement of tea tree genome sequencing is bringing tea tree biology out of the dark that will greatly help worldwide tea breeders to breed new varieties with more diverse tea taste without pesticide residues and also help to potentiate medicinal uses," said GAO, who called himself "a good tea drinker" for a long time.
"It is our hope that more new tea tree cultivars would finally satisfy and attract more tea drinkers worldwide." (Xinhua)

Camellia sinensis (Image by KIB)
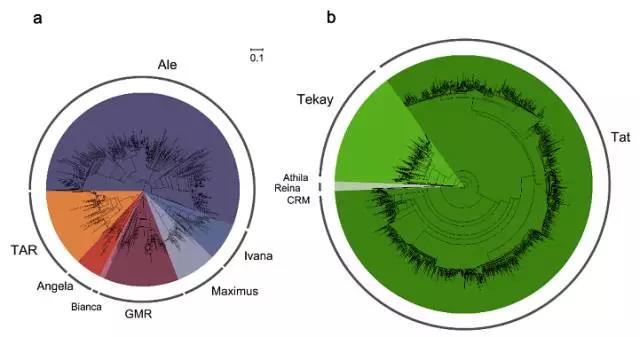
Fig1: Phylogenetic Analysis of the LTR Retrotransposon Sequences in the Tea Tree Genome (Image by KIB)
Contact:
Germplasm Bank of Wild Species
Kunming Institute of Botany, Chinese Academy of Sciences
Prof. GAO Lizhi
Email: lgao@mail.kib.ac.cn
(Editor:YANG Mei)




