BEP clade of the grass family (Poaceae) is composed of three subfamilies, i.e. Bambusoideae, Ehrhartoideae, and Pooideae. Controversies on the phylogenetic relationships among three subfamilies still persist in spite of great efforts. However, previous evidence was mainly provided from plastid genes with only a few nuclear genes utilized. Given different evolutionary histories recorded by plastid and nuclear genes, it is indispensable to uncover their relationships based on nuclear genes.
Recently, staff at Kunming Institute of Botany, headed by Profs. GUO Zhenhua and Prof. LI Dezhu and collaborators, have deeply investigated on this problem. Eleven species with whole-sequenced genome and six species with transcriptomic data were included in this study. A total of 121 one-to-one orthologous groups (OGs) were identified and phylogenetic trees were reconstructed by different tree-building methods. Genes which might have undergone positive selection and played important roles in adaptive evolution were also investigated from 314 and 173 one-to-one OGs in two bamboo species and 14 grass species, respectively. Our results support the ((B, P) E) topology with high supporting values. Besides, our findings also indicate that 24 and nine orthologs with statistically significant evidence of positive selection are mainly involved in abiotic and biotic stress response, reproduction and development, plant metabolism and enzyme etc., for example, WCOR413,PM5, Meu13, OsClp8, RRM and DNA-directed RNA polymerase II from two bamboo species and 14 grass species, respectively.
The results confirm that BEP clade is a monophyletic group and Bambusoideae is sister to Pooideae rather to Ehrhartoideae, which is in consensus with recent chloroplast-based phylogenomic trees. Meanwhile, these genes provide valuable insights into adaptive selection of the grass family at the sequence level and will be great candidates for future functional validation.
This work has been published online in PLoS ONE [Lei Zhao, Ning Zhang, Peng-Fei Ma, Qi Liu, De-Zhu Li, Zhen-Hua Guo. (2013). Phylogenomic Analyses of Nuclear Genes Reveal the Evolutionary Relationships within the BEP Clade and the Evidence of Positive Selection in Poaceae. PLoS ONE 8(5): e64642. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0064642]. The study was highly appreciated – “It has many merits. Otherwise, the idea and science behind your manuscript is very interesting” by Dr. Axel Janke (Academic Editor).
This study was supported by the Knowledge Innovation Project of the Chinese Academy of Sciences (KSCX2-YW-N-067); the National Natural Science Foundation of China (30990244); NSFC-Yunnan province joint foundation (U1136603); Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry and the Young Academic and Technical Leader Raising Foundation of Yunnan Province (No. 2008PY065, awarded to Zhen-Hua Guo).
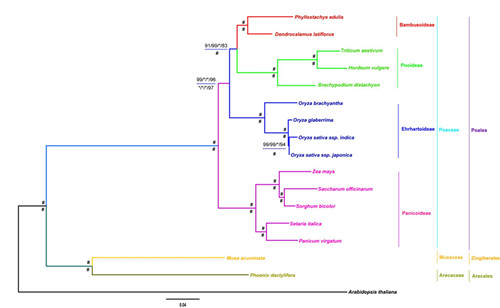
Phylogenetic relationships of the BEP Clade. Phylogenomic trees were inferred by the concatenation analyses, PAUP, RAxML and MrBayes.Speciestrees were also estimated by the coalescent method, MP-EST. The bootstrap values above the horizontal are based on protein, while the values below are based on nucleotide data.
“*” indicates support values of posterior probabilities (PP) = 1.0 and bootstrap (BP) = 100. “#” indicates all support values of PP = 1.0 and BP = 100. Support values are shown for nodes as maximum parsimony bootstrap/maximum likelihood bootstrap/Bayesian inference posterior probability/maximum pseudo-likelihood model bootstrap. Branch lengths were estimated through protein super-matrix using Bayesian analysis, and scale bar denotes substitutions per site.




