New genes are critical for speciation, evolution and adaptation of species. Horizontal gene transfer (HGT) is an important way of acquiring new genes and its occurring scales and biological significance have been extensively investigated in prokaryotes and unicellular eukaryotes. In higher plants, HGT events have been found mainly between the organelle genomes from different plants and were rarely reported between the nuclear genomes of plants.
Members from the groups of Dr. WU Jianqiang, Dr. HUANG Jinling, Dr. LI Suoping, and Dr. WEN Jianfan reported a newly evolved strictosidine synthase-like gene (SSL) in Brassicaceae was co-opted by two parasitic plants, the root parasitic plant Orobanche aegyptiaca and the stem parasitic plant Cuscuta australis, by independent HGT events. These findings were strongly supported by phylogenetic analysis and the identical unique amino acid residues and deletions of SSL sequences in the parasitic plants and in Brassicaceae. The presence of introns indicated that the transfer occurred directly by DNA integration in both parasites. Furthermore, positive selection was detected in the foreign SSL gene in O. aegyptiaca but not in C. australis. The expression of the foreign SSL genes in these two parasitic plants was detected in multiple development stages and tissues, and the foreign SSL gene was induced after wounding treatment in C. australis stems. These data imply that the foreign genes may still retain certain functions in the recipient species. The study suggests a potential role of the horizontally transferred genes in the evolution and adaptation to parasitic lifestyle or environment (http://www.biomedcentral.com/1471-2229/14/19/abstract).
This work was supported by the grants from NSFC (No. 31301037) and NSF of Yunnan Province of China (No. 2013FB068) to Dr. Guiling Sun and the grants of Yunnan Recruitment Program of Experts in Sciences (No. 2012HA016) and the Thousand Youth Talents Program to Dr. Jianqiang Wu.

Figure 1. Molecular phylogeny and partial alignment of the Brassicaceae-specific SSL proteins and the foreign SSLs in O. aegyptiaca and C. australis. Sequence IDs are shown after the species names. Numbers above or below the branches indicate bootstrap support values of maximum likelihood no less than 75%. The sequences from parasitic plants O. aegyptiaca and C. australis are highlighted in boxes. The respective partial protein sequences are shown on the right side of the species names and sequence IDs. The amino acids and the single amino acid deletion uniquely conserved in Group II are shaded.
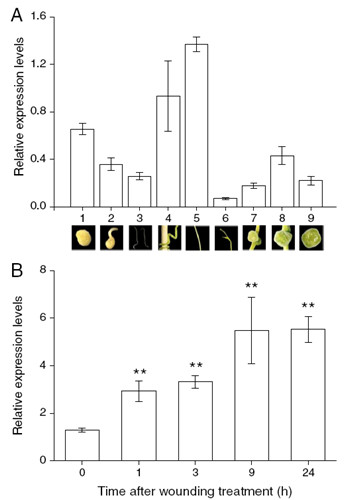
Figure 2. Expression level analyses of the CaSSL gene in C. australis during different developmental stages and after wounding treatment by qRT-PCR. A. Expression levels (±SE, n=5) of the CaSSL gene at different developmental stages and in different organs. 1, seeds; 2, just germinated seeds; 3, seedlings; 4, pre-haustoria; 5, stems; 6, shoot tips; 7, floral buds; 8, flowers; 9, capsules. A picture of each developmental stage was shown below its expression labeling. B. Expression levels (±SE, n=5) of the CaSSL gene after wounding treatment in stems. Asterisks indicate significant differences between treatment and 0-h time point (** P < 0.01; Student’s t-test).




