DNA methylation, one of important epigenetic markers, has shown broad-ranging functions, including regulation of gene expression and genomic imprinting, involvement in chromatin organization and protection of genome from invading and mobile DNA elements.
In plants, DNA methylation generally refers to three different nucleotide sequence contexts: symmetric CG and CHG methylation and asymmetric CHH methylation (where H = C, T or A). Generally, the CG methylation is maintained by the conserved DNA methyltransferase MET1, and the CHG methylation is maintained by the plant-specific DNA methyltransferase CMT3, whereas CHH de novo methylation is established by 24-nt siRNA-directed DNA methylation (RdDM) pathway to guide the DNA methyltransferase DRM1/2 in plants. It has been known that active DNA demethylation mainly, in Arabidospsis, depends upon activity of the DNA glycosylase including DME, ROS1, DML2/3, which is able to recognize and remove methylated cytosines by a base excision-repair pathway. Clearly, the extents and patterns of genomic DNA methylation are usually maintained by the interaction of DNA methyltransferases and demethylases.
Increasing evidences demonstrate that DNA methylation plays crucial roles in the control of seed development and seed size. Investigations on genomic DNA methylation in the seeds of Arabidopsis, rice and maize revealed extensive hypo-methylation in the endosperm genome. However, the mechanism about how the extent and pattern of DNA methylation is established and whether the extent and pattern of DNA methylation is conserved in plant seeds are largely unclear.
Specifically, in most dicots including Arabidopsis their endosperms are ephemeral and gradually consumed by the embryo tissue during seed development. Due to this reason it appears difficult to dissect the potential mechanism of genomic DNA methylation occurring in dicot’s seeds. In contrast to Arabidopsis, castor bean (Ricinus communis L.), a member of Euphorbiaceae family, has relatively large and persistent endosperm throughout seed development, which provides an ideal model plant to dissect the potential mechanism underlying seed genomic DNA methylation arises and their biological interests with seed development in dicots.
Very recent news, Prof. LIU Aizhong and his colleagues at the Kunming Institute of Botany, Chinese Academy of Sciences (CAS) comprehensively dissected the genomic DNA methylation in castor bean seeds. Unexpectedly, they discovered a novel genomic DNA methylation profile that the CHH methylation extents in endosperm and embryo were substantially higher than known plants, irrespective of the CHH percentage in their genomes.
Very interestingly, they revealed that the endosperm exhibited a global reduction in the CG and CHG methylations relative to the embryo, markedly switching the global gene expression, however, the CHH methylation occurring in endosperm did not exhibit a significant reduction. Combining with the expression of 24-nt siRNAs mapped within TE regions and genes involved in the RdDM pathway, they demonstrated that the 24-nt siRNAs played a critical role in maintaining CHH methylation and repressing the activation of TEs in endosperm persistent development.
In short, their study exhibited a novel genomic DNA methylation profile occurring in dicot seeds with persistent endosperm, which gives new insights into understanding the genomic DNA methylation arising and its biological interests in plants.
This study has been published online in Plant physiology: http://www.plantphysiol.org/content/early/2016/04/28/pp.16.00056.1.full.pdf+html
Authors are grateful for the financial support from the Chinese National Key Technology R & D Program (2015BAD15B02), National Key Basic Research Program of China (2014CB954100) and the National Natural Science Foundation of China (31501034).
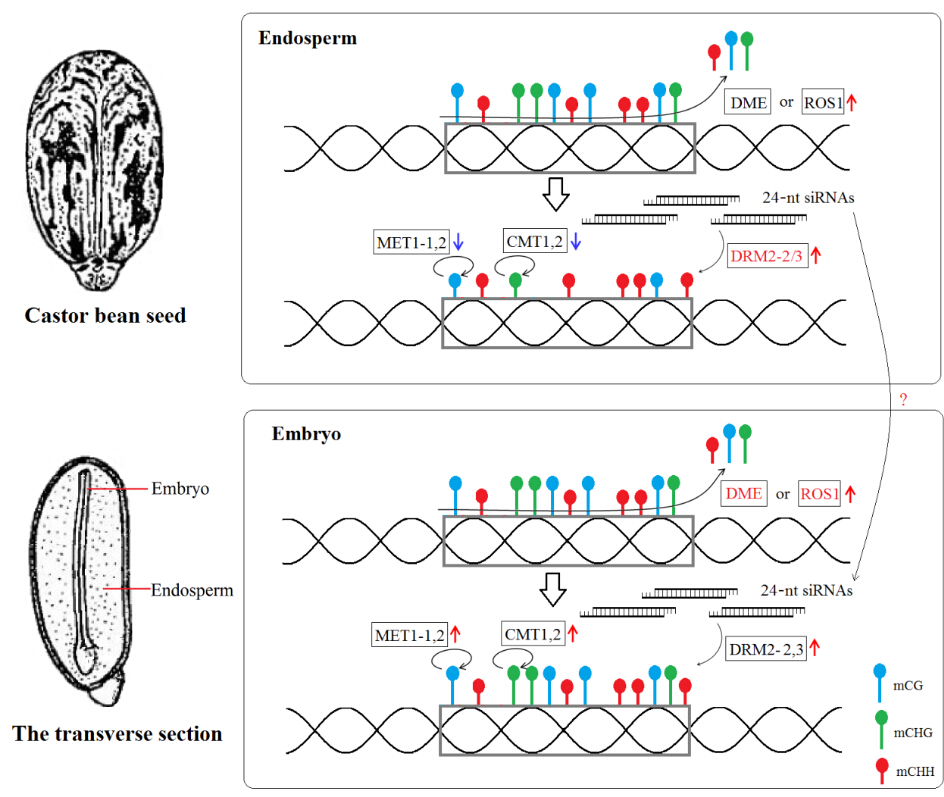
Figure: A proposed genomic DNA methylation pattern
Contact for this research: LIU Aizhong or XU Wei
Department of Economic Plants and Biotechnology
Yunnan Key Laboratory for Wild Plant Resources
Kunming Institute of Botany, Chinese Academy of Sciences
Email: liuaizhong@mail.kib.ac.cn, or xuwei@mail.kib.ac.cn




