ddRAD-seq is a reduced representation sequencing technology by sampling genome-wide restriction-site enzyme loci developed on the basis of next-generation sequencing. This technique has been widely applied to SNP marker development and genotyping on animals, especially on marine metazoa as the original ddRAD protocol was mainly built and trained based on animal data. However, wide application of ddRAD-seq technology in plant species has not been achieved so far. It is necessary to develop an optimized ddRAD library preparation protocol accessible to most angiosperm species without much startup pre-experiment and lower cost.
Researchers at Prof. GUO Zhenhua and Prof. LI Dezhu’ s group of Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS), in collaboration with Prof. LI Li from theInstitute of Oceanology, Chinese Academy of Sciences (IOCAS) first tested several combinations of enzymes by in silico analysis of 23 plant species (covering 17 families of angiosperms and 1 family of bryophyte) and found AvaII + MspI enzyme pair produced consistently higher number of fragments in a broad range of plant species.
Then the research team removed two purifying and one quantifying steps of the original protocol, replaced expensive consumables and apparatus by conventional laboratory apparatus. Besides, the team shortened P1 adapter from 37bp to 25bp and designed a new barcode-adapter system containing 20 pairs of barcodes of varying length. The optimized ddRAD strategy for angiosperm plants is economical, time-saving and requires little technical expertise or investment in laboratory equipment. This simplified protocol was referred to MiddRAD by Dr. YANG Guoqian, who made major contributions to this approach and demonstrated the utility and flexibility of this approach in resolving phylogenetic relationships of two genera of woody bamboos (Dendrocalamus and Phyllostachys). Their optimization of ddRAD may make it widely used in fields of plant population genetics, phylogenetics, phylogeography and molecular breeding.
A paper entitled “Development of a universal and simplified ddRAD library preparation approach for SNP discovery and genotyping in angiosperm plants” has recently been published in Plant Methods: http://plantmethods.biomedcentral.com/articles/10.1186/s13007-016-0139-1 .
This study was supported by Natural Science Foundation of China (Grant No. 31470322 and 31430011).
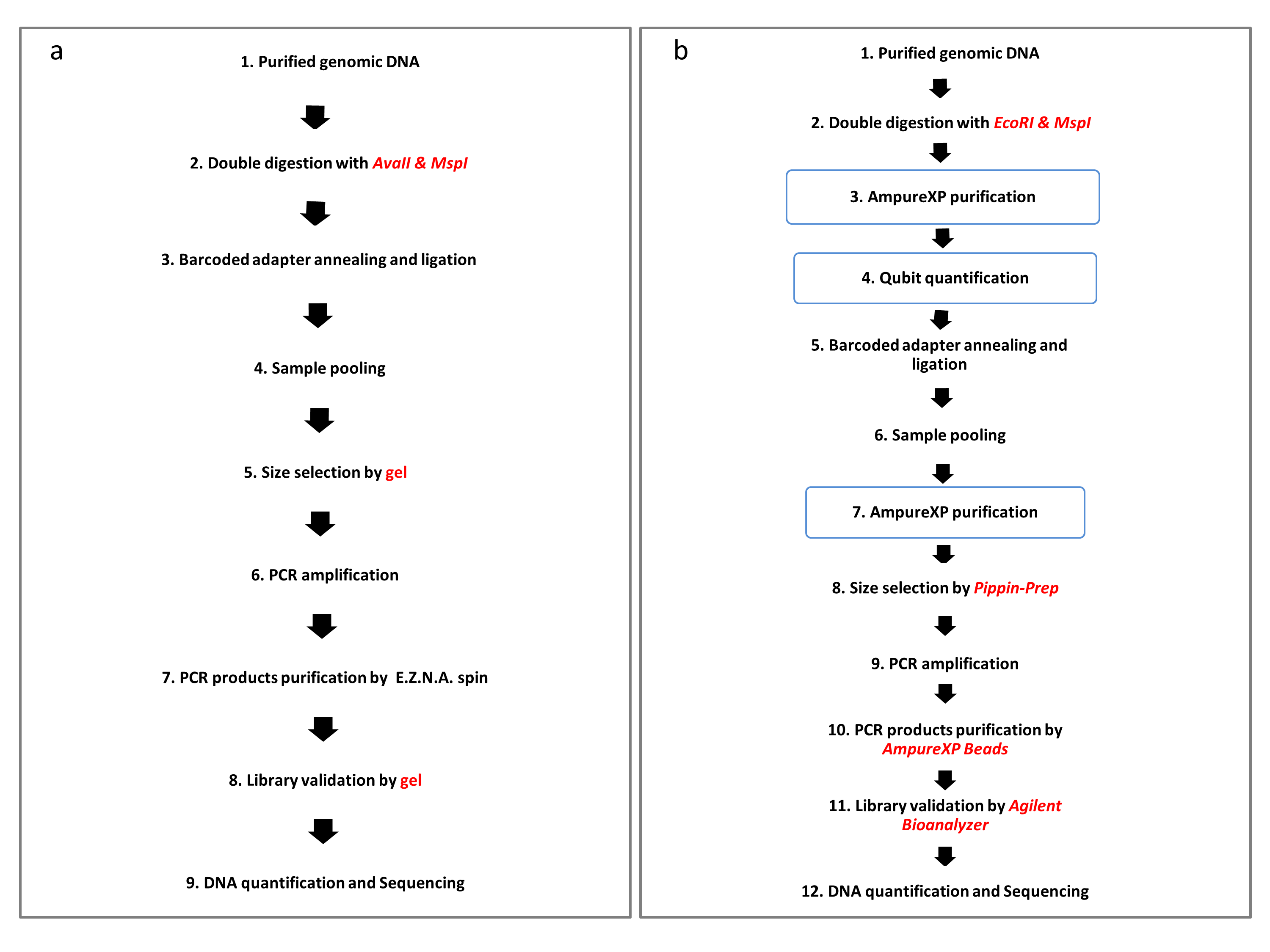
Fig. 1: Library preparation flowchart of MiddRAD protocol B and the original ddRAD protocol. a MiddRAD protocol B. b The original ddRAD library constructing flowchart, this procedure is adopted by Peterson et al. Protocol B contains nine steps while the original ddRAD protocol contains 12 steps.
Contact:
Germplasm Bank of Wild Species,
Kunming Institute of Botany, Chinese Academy of Sciences
Prof. GUO Zhenhua & Prof. LI Dezhu
Email: guozhenhau@mail.kib.ac.cn, dzl@mail.kib.ac.cn




