Phylogenomics applies genomic data to reconstruct the evolutionary history of organisms and has been applied to tackle rapidly radiating clades and hybridization events in diverse lineages. Due to their moderate size, moderate nucleotide substitution rates, and freedom from problems of paralogy, plastid DNA sequences have been widely used for the reconstruction of plant phylogenies.
Rosaceae are one of the most diverse angiosperm families with ca. 90 genera and 3000 species with a global distribution. Many species of Rosaceae are economically important, either as edible fruits, ornamentals, and timbers. As an ecologically and economically important group, Rosaceae have been a special focus of many botanists, and a series of taxonomic and phylogenetic studies have been published. However, portions of the tree remained unresolved, with weakly supported and conflicting relationships. And, the large-scale evolutionary history of Rosaceae remains poorly understood, although previous dating analyses have provided some insights.
Researchers from Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS) analyzed 130 newly sequenced plastomes together with 12 from GenBank of Rosales in an attempt to reconstruct deep relationships and reveal temporal diversification of this family.
The results highlight the importance of improving sequence alignment and the use of appropriate substitution models in plastid phylogenomics. Three subfamilies and 16 tribes (as previously delimited) were strongly supported as monophyletic, and their relationships were fully resolved and strongly supported at most nodes.
Rosaceae were estimated to have originated during the Late Cretaceous with evidence for rapid diversification events during several geological periods. The major lineages rapidly diversified in warm and wet habits during the Late Cretaceous, and the rapid diversification of genera from the early Oligocene onwards occurred in colder and dryer environments. The robust phylogenetic backbone and time estimates provided in this study established a framework for future comparative studies on rosaceous evolution.
The research team includes groups led by Dr. LI Dezhu and Dr. YI Tingshuang from the KIB/CAS, Mark W. Chase form Royal Botanic Gardens, Kew and Douglas E. Soltis from Florida Museum of Natural History, University of Florida participated in and made contributions to this work.
This study was supported by the grants from the National Key Basic Research Programme of China (2014CB954100), the Sino-Africa Joint Research Center (SAJC201302), the National Science and Technology on Basic Research Programme (2013FY112600), the Science and Technology Innovation of CAS, iFlora Cross and Cooperation team (31129001), and the Talent Project of Yunnan Province (2011CI042).
This study was facilitated by the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences.
The research entitled “Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics” has been published in New Phytologist. (http://onlinelibrary.wiley.com/doi/10.1111/nph.14461/full)
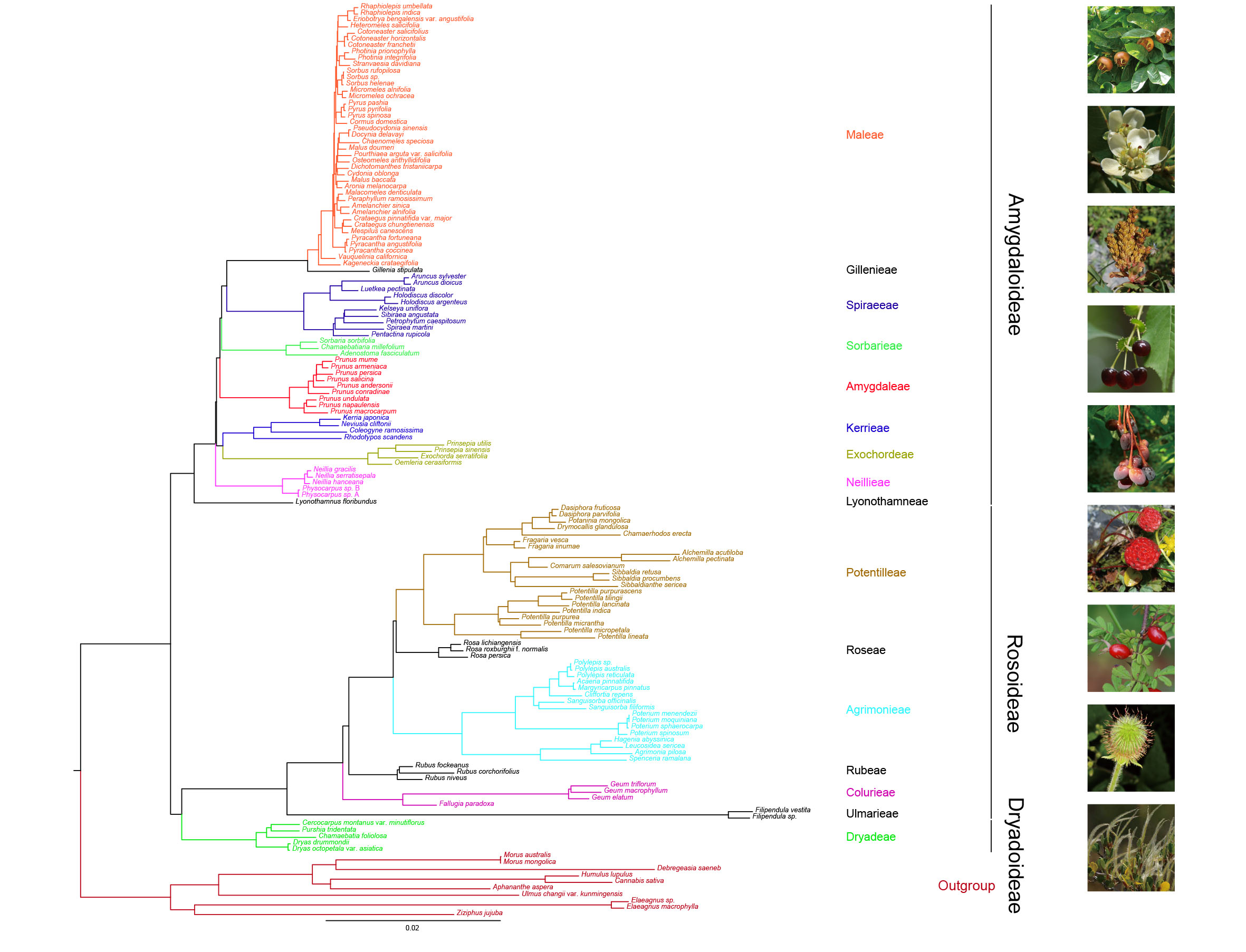
Fig.1 The ML tree of Rosaceae. (Image by KIB)
Contact:
Plant Germplasm and Genomics Center
Germplasm Bank of Wild Species
Kunming Institute of Botany, Chinese Academy of Sciences
Prof. Dr. YI Tingshuang
Email: tingshuangyi@mail.kib.ac.cn




