Species identification is the keystone of biodiversity conservation. For the endangered and economically important IUCN- and CITES- listed taxa, a rapid and accurate species identification system is a critical component of bio-surveillance, conservation management, and potentially policing illegal trades, which in turn will mitigate the extinction risk of biodiversity.
It is hardly possible to use traditional taxonomy to identify fragmented samples or pre-processed plants to species.
Reliable identification can be achieved via a comprehensive DNA barcode reference library, accompanied by precise geographical distribution data. However, these require extensive sampling at spatial and taxonomic scales, which has rarely been achieved for cosmopolitan taxa.
Taxus is the most diverse genus within Taxaceae, and it is broadly distributed across temperate of the Northern Hemisphere.
It has acquired great medical significance as the natural source of taxol, an anti-tumor agent with high potential for cancer treatments.
Species of Taxus are slow-growing and scatteredly distributed, thus rarely occurring in large numbers.
Consequently, commercial exploitation and the illegal trade of its bark and leaves for taxol has caused a sharp decline in its natural populations.
According to IUCN, five species under the previous scheme of classification are classified to the category “Endangered” and four are listed in CITES Appendix II.
At the national level, all native Chinese Taxus species are listed as first-class national protected plants.
Due to the taxonomic difficulty, the genetic and geographical boundary among species is largely uncertain, an accurate, quick, cost-effective and universally applicable identification system for Taxus species is urgently needed to support and enforce international and national plant protection laws.
Recently, a collaborative research team led Prof. GAO Lianming and Prof. LI Dezhu from Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS), constructed a reliable and repeatable identification system by combing a comprehensive DNA barcode reference library and a precise geographical distribution map for all recognized 15 yew species worldwide.
The system includes the DNA and distribution ID for each species of Taxus, and was successfully applied in three forensic identification cases.
The work set a reproducible exemplar for other IUCN- and CITES-listed taxa.
The results suggested that trnL-trnF is the ideal barcode for Taxus: it can distinguish all Taxus species, and in combination with ITS to identify hybrids.
Among five analysis methods tested, NJ was the most effective method for species identification. By screening 4151 individuals for trnL-trnF from 251 populations of 15 species, 73 haplotypes were detected, all of them species-specific and some population private.
Taxonomical, geographical and genetic dimensions of sampling strategy were all together found to affect the comprehensiveness of the resulting DNA barcode library. Maps from species distribution modelling (SDM) showed that most species had allopatric distributions, except T. mairei in the Sino-Himalayan region.
Using the DNA barcode library and distribution map, two unknown forensic samples were identified to species (while one sample identified to geographic origin), and another was determined as a putative interspecific hybrid.
This system can determine both species and hybrids, and can in some cases greatly narrow down the geographical location.
This integrated species identification system for yews can be used for bio-surveillance, conservation management and to monitor and prosecute illegal trade.
The work will serve as an effective tool for species identification for IUCN- and CITES-listed species.
This will in turn reinforce the objectives of international treaties, strengthen national forestry management, and help enforce conservation laws designed to curb the increasing threat of illegal exploration and illicit trade, all of which will partly mitigate the extinction risk of species.
The study entitled “Integrating a comprehensive DNA barcode reference library with a global map of yews (Taxus L.) for forensic identification” has been published online on “International Day for Biological Diversity” (22 May) in Molecular Ecology Resources.
This work was supported by the National Key Basic Research Program of China, National Natural Science Foundation of China and Interdisciplinary Research Project of KIB.
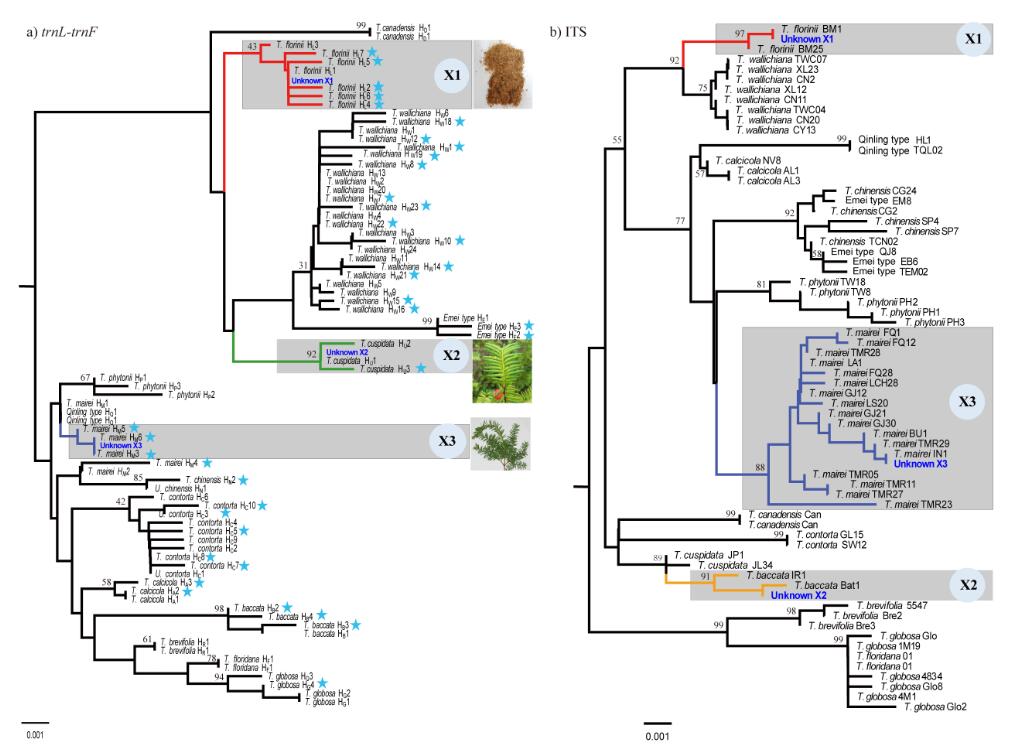
Fig. 1 The clustering relationship of three unknown samples in the trnL-trnF (a) and ITS (b) NJ tree of reference haplotypes. The cyan stars in fig. (a) indicate private trnL-trnF haplotypes. Clades that include “Unknowns” are highlighted with gray squares, X1, X2 and X3 represent the three unknown samples. Bootstrap values are shown on the branches. (Image by KIB)
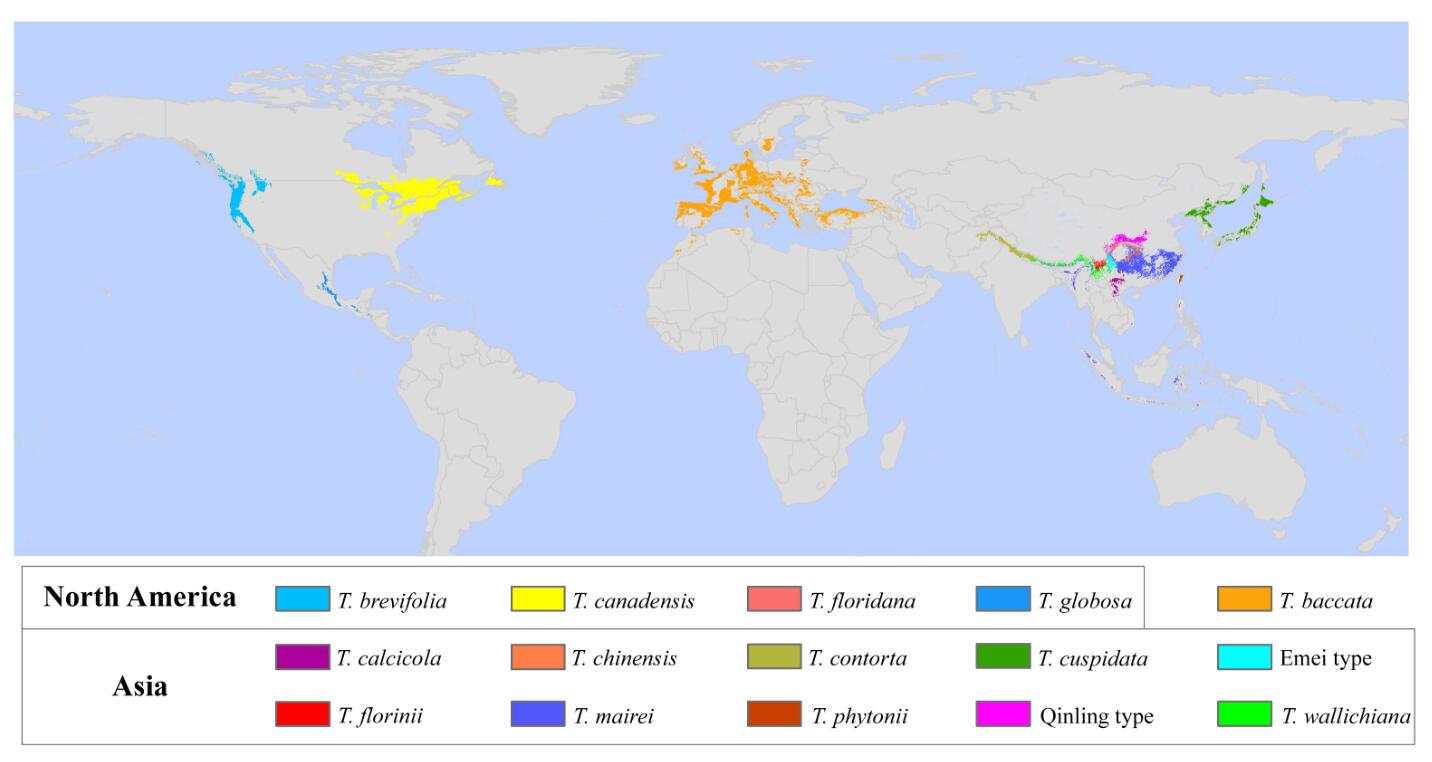
Fig. 2 The potential global geographical distribution of 15 Taxus species predicted using species distribution modelling. (Image by KIB)
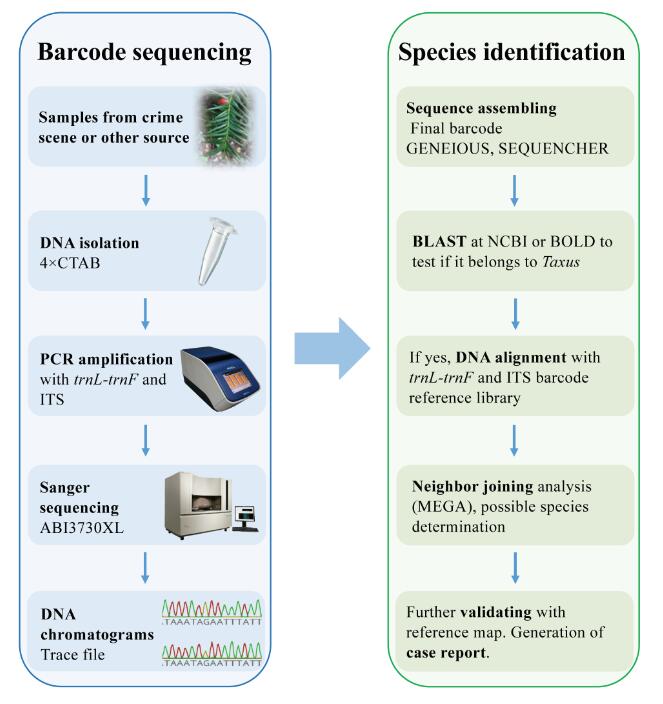
Fig. 3 The applicable workflow for species identification of unknown Taxus samples using the DNA barcode libraries and reference map generated in this study. (Image by KIB)
Contact:
Yang Mei
General office
Kunming Institute of Botany, CAS,
Email: yangmei@mail.kib.ac.cn
(Editor:YANG Mei)




