The grass family (Poaceae) is one of the largest families within angiosperms with about 12,000 species, providing with most of food crops for human and domestic animals as well as other extensive utilities.
The grass family is also a long-standing model system for genetic and genomic research of plants. Among 12 subfamilies of the grass family, Bambusoideae (bamboos), along with the Pooideae (e.g., Brachypodium and wheat), the Oryzoideae (e.g., rice) comprise the BOP clade and is the only major grass lineage that diversified in mesic to wet forests.
Bamboos are among the fastest growing plants on earth and the tree-like woody grasses rarely flower, with a vegetative phase up to 120 years. As a peculiar subfamily of grasses, there has been a black box of knowledge for bamboos regarding chromosomal and genomic evolution due to their particularly long life-cycle and unpredictable flowering.
The nearly 1,500 described bamboo species fall into four monophyletic lineages with different ploidy levels according to the basic chromosome number of bamboos as 12: herbaceous bamboos (HB, 2n=20-24) are mostly diploids, temperate woody bamboos (TWB, 2n=46-48) and neotropical woody bamboos (NTB, 2n=40-46) are tetraploids, while paleotropical woody bamboos (PTB, 2n=70-72) are hexaploids.
Nevertheless, both the origins of the woody bamboos and their relationships to herbaceous lineage remain uncertain. Understanding of their unique biological traits is also far from resolved.
A research group lead by Prof. LI Dezhu from Kunming institute of Botany, Chinese Academy of Sciences (KIB/CAS), has long focused on research for taxonomy, phylogenetics and evolution of bamboos. Collaborated with Institute of Plant Physiology and Ecology, Chinese Academy of Sciences and Shanghai Ocean University, the team recently accomplished the study of “web of life” and comparative genomics of unique traits of bamboos based on the whole genome sequencing, facilitated by a national large-scale scientific facility, the Germplasm Bank of Wild Species.
This study generated four draft genomes from Olyra latifolia (Ola) and Raddia guianensis (Rgu) from herbaceous, Guadua angustifolia (Gan) from neotropical (NTB), and Bonia amplexicaulis (Bam) from paleotropical (PTB) bamboos (Fig.1), representing diploid, tetraploid and hexaploid as described above. Two high-quality assemblies were constructed for B. amplexicaulis and O. latifolia.
The team also constructed the first high-density genetic linkage map of bamboo for a hexaploid species, Dendrocalamus latiflorus. Genome-wide gene synteny analyses confirmed the diploid origin of herbaceous and polyploidy of woody bamboos, consistent with the previous cytological results.
To address the genome evolution of polyploid bamboos more deeply, this research explored a new phylogenomic approach based on syntenic genes with ‘perfectly’ expected copies based on subgenome characterization and ploidy levels. Together with the published moso bamboo (Phyllostachys edulis, Ped) genome, comparative genomics was conducted for total five bamboo genomes and revealed a complex reticulate evolution of bamboos.
In the results, herbaceous bamboo lineage was diverged from the progenitor genomes of woody bamboos at ca. 42 million-years ago (Ma). The extant three clades of woody bamboos originated from four extinct diploid ancestors (designated as A, B, C, and D) by three independent allopolyploid events at ca. 22 Ma.
The ancestral B+C and C+D genomes hybridized to generate two allotetraploid lineages, one represented by the NWB clade (G. angustifolia, BBCC) in the tropics and another by the TWB clade in temperate or montane areas (P. edulis, CCDD). A third hybridization occurred between the ancestral A and B+C genomes at around 19 Ma in the early Miocene, leading to the origin of the allohexaploid PWB clade (B. amplexicaulis, AABBCC) (Fig. 2).
This could have been followed by the onset and intensification of the East Asian Monsoon to facilitate the successful adaptation of woody bamboos to the mesic to wet forests. The C subgenome is shared by all the extant woody bamboo lineages and, based on the genetic map, has undergone more intra-chromosome inversions and rearrangements relative to A, B and D subgenomes.
Therefore, the progenitor of the C subgenome could have taken adaptive advantages and the polyploids harboring it have radiated successfully. In addition, comparative analyses on flowering genes found that lost-/mal-function of SOC1-like genes, positively selected OsPRR95-like genes, and copy number variation of GA pathway enzymes associated together to regulate the long vegetative growth in woody bamboos (Fig. 3).
As a successful case, this study can serve as a potential model for furthering genome research on origin and complicated evolutionary scenarios of other angiosperm lineages, and will greatly enhance research on bamboo biology.
The paper was recently published online in Molecular Plant entitled 'Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos'.
This work was supported by the Strategic Priority Program of the CAS (XDB31000000), the National Natural Science Foundation of China (grants 31430011 and 31670227), Leading Talents Program of Yunnan Province (2017HA014), CAS Youth Innovation Promotion Association (2015321), The grant from Germplasm Bank of Wild Species (Y77P4412Z1), and CAS Pioneer Hundred Talents Program (292015312D11035).
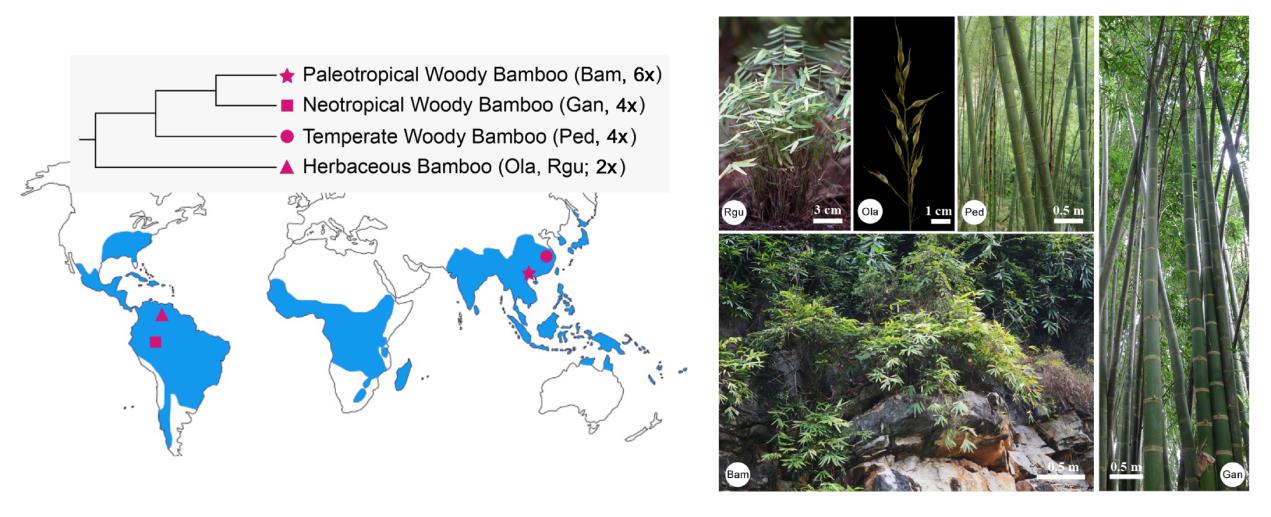
Figure 1. Sampling of the five bamboos and the representative major clades (refer to the main text for species acronyms). (Image by KIB)
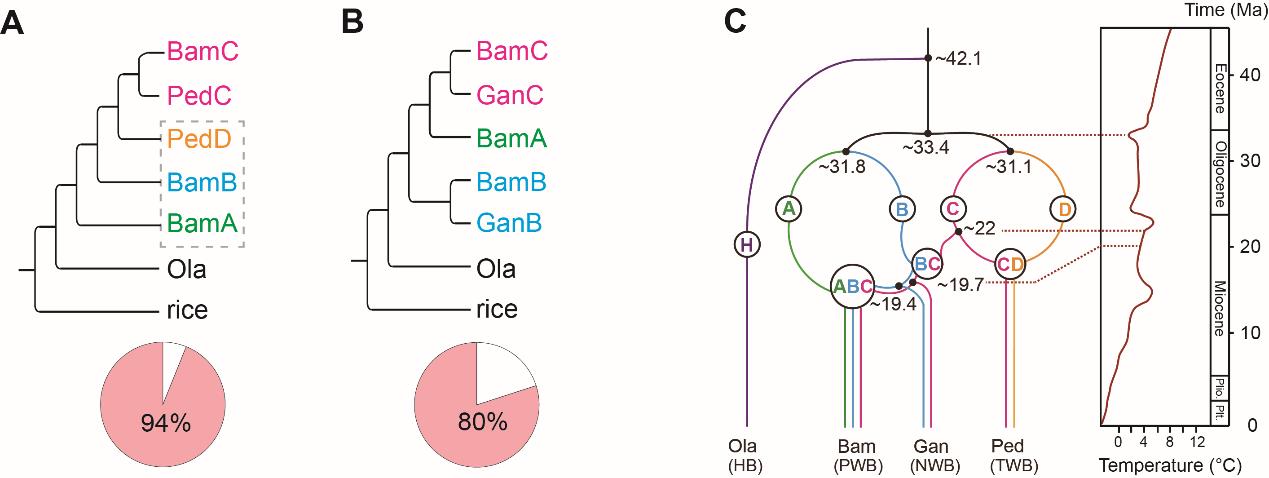
Figure 2. The allopolyploidization of woody bamboos (refer to the main text for species acronyms; Ma: million-year ago). (Image by KIB)
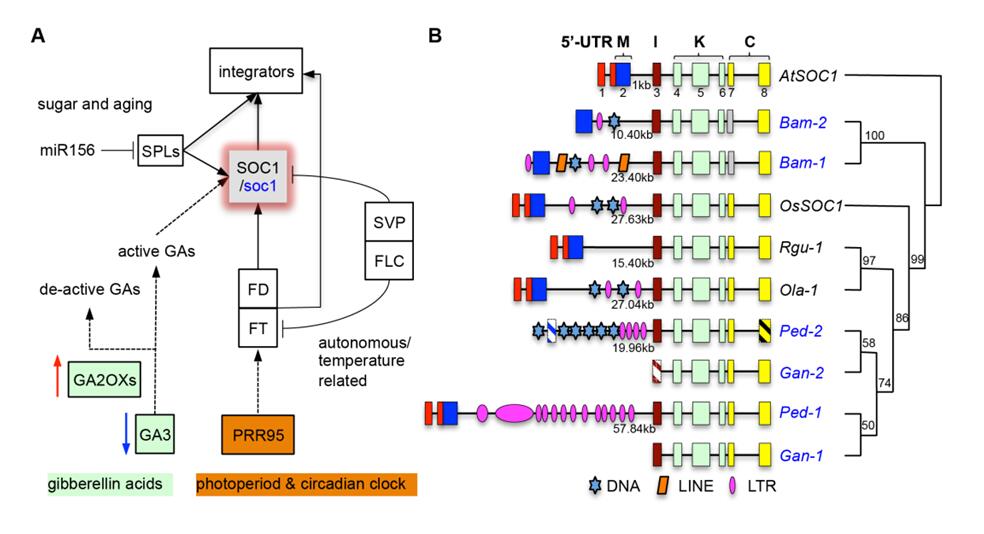
Figure 3. The flowering network of woody bamboos (refer to the main text for species acronyms). (Image by KIB)
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
Email: yangmei@mail.kib.ac.cn
(Editor: YANG Mei)




