Leguminosae (also known as Fabaceae) are the third largest angiosperm family, containing over 19,500 species in ca. 765 genera, as well as one of the most economically important families.
Legumes comprise a huge diversity of morphologies and habits, including tiny annual herbs, vines, shrubs, lianas and long-lived trees.
Legumes are widely distributed around the world, and about 88% of them have the ability to establish associations with nitrogen-fixing rhizobia and hence play an important role in the maintenance of sustainable agriculture and ecosystem function.
Legumes are used in many aspects of human life, including as foods, oils, fodders, woods, medicines, textiles, biological products (such as industrial enzymes), ornamental and horticultural plants.
Dozens of legumes have been domesticated and cultivated as major crops that provide highly nutritious sources of protein. In some rural areas, legumes are even the only source of protein for human consumption.
During the past two decades, considerable progress has been made in resolving phylogenetic relationships using molecular data.
In 2017, the Legume Phylogeny Working Group proposed a new six-subfamily classification system, based on a well sampled legume phylogenetic tree inferred from the gene matK, and combined with morphological evidence. However, previous studies have failed to resolve many important relationships with strong support, in part due to the small number of included gene fragments and/or the limited sampling of most previous studies. Consequently, a robust phylogenetic framework has been lacking for this globally important group.
Basing on the national, large-scale scientific facility of the Germplasm Bank of Wild Species, YI Tingshuang’s and LI Dezhu Li’s groups from Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS), used 187 representative species (151 of them were newly sequenced) from six subfamilies and 97% of the recognized tribes of Leguminosae, and from the other three families of Fabales to reconstruct a comprehensive phylogeny for Leguminosae.
Most analyses of the coding regions (PC), noncoding regions (PN), and the combined coding and noncoding regions (PCN) (including 3 basic datasets, 23 derived datasets and 6 datasets derived from phylogenetic signal analyses) produced largely congruent trees with strong statistical support, thus providing strong support for resolution of some long-controversial deep relationships among the early diverging lineages of the subfamilies Caesalpinoideae and Papilionoideae (Fig. 1).
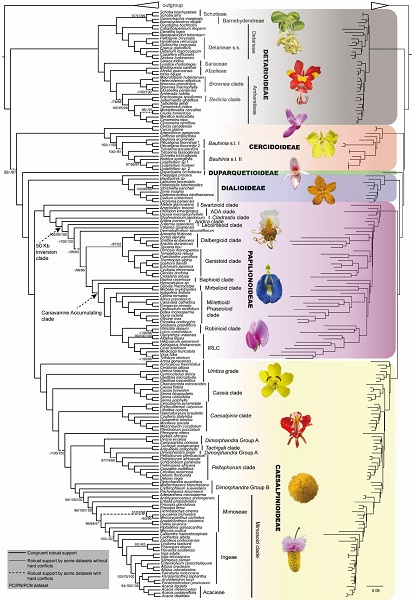
Fig. 1 The maximum-likelihood tree of Leguminosae derived from the plastid phylogenomic analysis of a concatenated dataset including coding and noncoding regions (PCN dataset).(Image by KIB)
In addition, this study confirmed the existence of conflicting phylogenetic signals in the plastome through the use of phylogenetic signal detection and quantification of several key conflicting nodes (Fig. 2), and through assessments gene tree congruence.
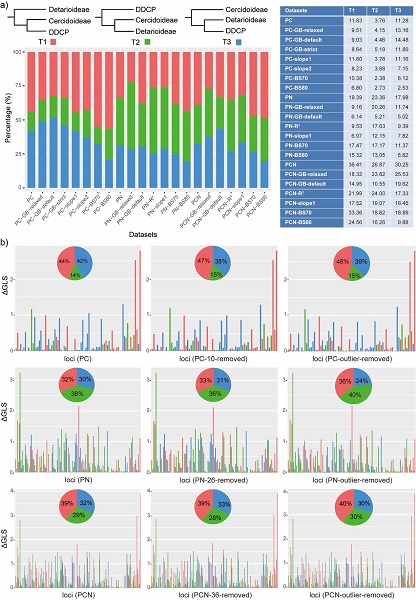
Fig. 2 The distribution of phylogenetic signal for three alternative topological hypotheses at the root of Leguminosae.(Image by KIB)
This study suggests the plastomes of many legumes might have complex evolutionary histories, perhaps involving phenomena such as biparental inheritance and heteroplasmic recombination.
If analyzing the entire plastome as a single evolutionary unit might result in misleading phylogenetic inferences, it is therefore necessary to thoroughly examine conflicting phylogenetic signal in plastid phylogenomics. The robust phylogenetic backbone reconstructed in this study establishes a framework for future studies on legume classification, evolution, and diversification.
With the title “Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae”, this study was recently published online in the top international evolutionary biology journal, Systematic Biology.
The work was a collaborative project by botanists from America, Australia, Brazil and Canada. The group members Dr. ZHANG Rong, Dr. WANG Yinhuan, and Dr. JIN Jianjun are co-first authors of the paper; Prof. YI Tingshuang and Prof. LI Dezhu are co-corresponding authors.
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31010000), the National Natural Science Foundation of China [key international (regional) cooperative research project No. 31720103903], and the Large-scale Scientific Facilities of the Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02).
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
email: yangmei@mail.kib.ac.cn
(Editor:Yang Mei)




