The plastid genome (plastome, plastids including the chloroplast and other plastid forms) and mitochondrial genome (mitogenome or chondriome) represent the portions of endosymbiotic organelle inheritance in eukaryotes that have remained in organelles without being transferred to the nucleus or lost.
The DNA sequenes from the organelle genomes have been widely used in phylogenetic and evolutionary analyses, and DNA barcoding. Due to high copy numbers of the organelle genome in a single cell, it is feasible to get enough coverage from the low coverage whole genome sequencing (WGS) data to assemble complete organelle genomes.
With the rapid advances of high throughput sequencing technologies, a tremendous amount of WGS data were produced in low cost, which makes the accurate and high throughput assembly of organelle genomes in great need. Although many toolkits and pipelines for assembling organelle genomes have been developed, their assembly qualities and efficiencies are generally below expectations.
Teams led Prof. LI Dezhu and Prof. YI Tingshuang from Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS) have been engaged in plastid phylogenomics, comparative genomics, and DNA barcoding for years.
Jointly, they have established a research system of utilizing the plastome data for phylogenetic and evolutionary studies, and achieved fruitful results (Ma et al., 2014.Systematic Biology;Zhang et al., 2017.New Phytologist;Li et al., 2019.Nature Plants;Zhang et al.,2020Systematic Biology).
Teams were also dedicated to develop new tool for plastome analyses, including a popular plastome annotation toolkit, PGA (Qu et al., 2019,Plant Methods).
Aiming at accurate and efficient organelle genome assembly, an international joint team led by Profs. Li and Yi, with collaborators from KIB and , Xishuangbanna Tropical Botanical Gardens, Chinese Academy of Sciences (XTBG/CAS), and Pennsylvania State University, newly developed GetOrganelle, an advanced toolkit for de novo assembling accurate organelle genomes.
The “from-reads-to-organelle” process, i.e. the main workflow of GetOrganelle can be summarized into the following five steps:
1) recruiting initial “seed reads” and estimating parameter,
2) recruiting more target-associated reads through extending,
3) conducting de novo assembly and generating assembly graph,
4) roughly filtering for target-like contigs,
5) identifying target contigs and exporting all configurations.
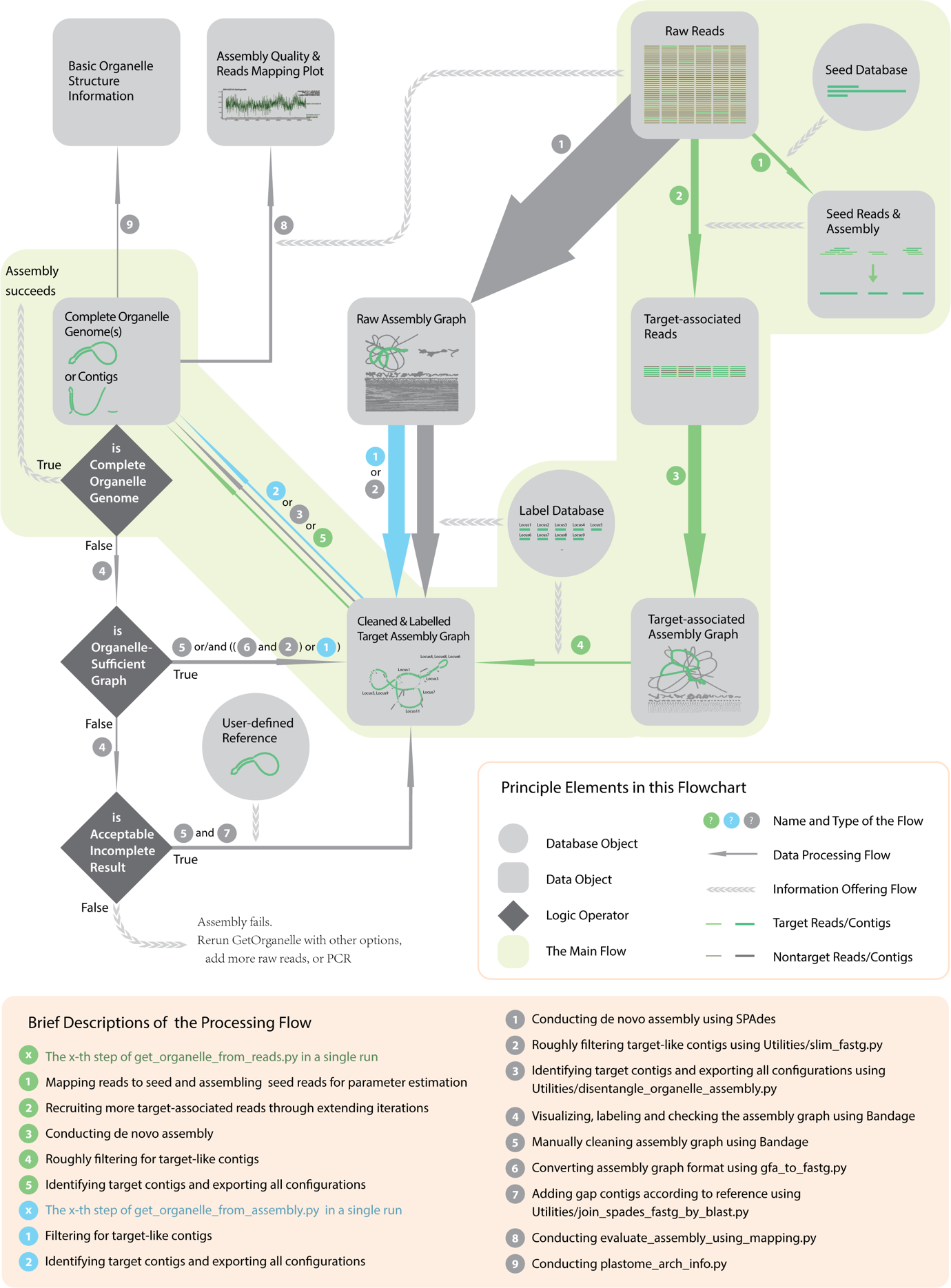
It is innovative that GetOrganelle provides a pre-grouping strategy for speeding up target-associated reads recruitment and contig multiplicity estimation algorithm for better repeat resolution.
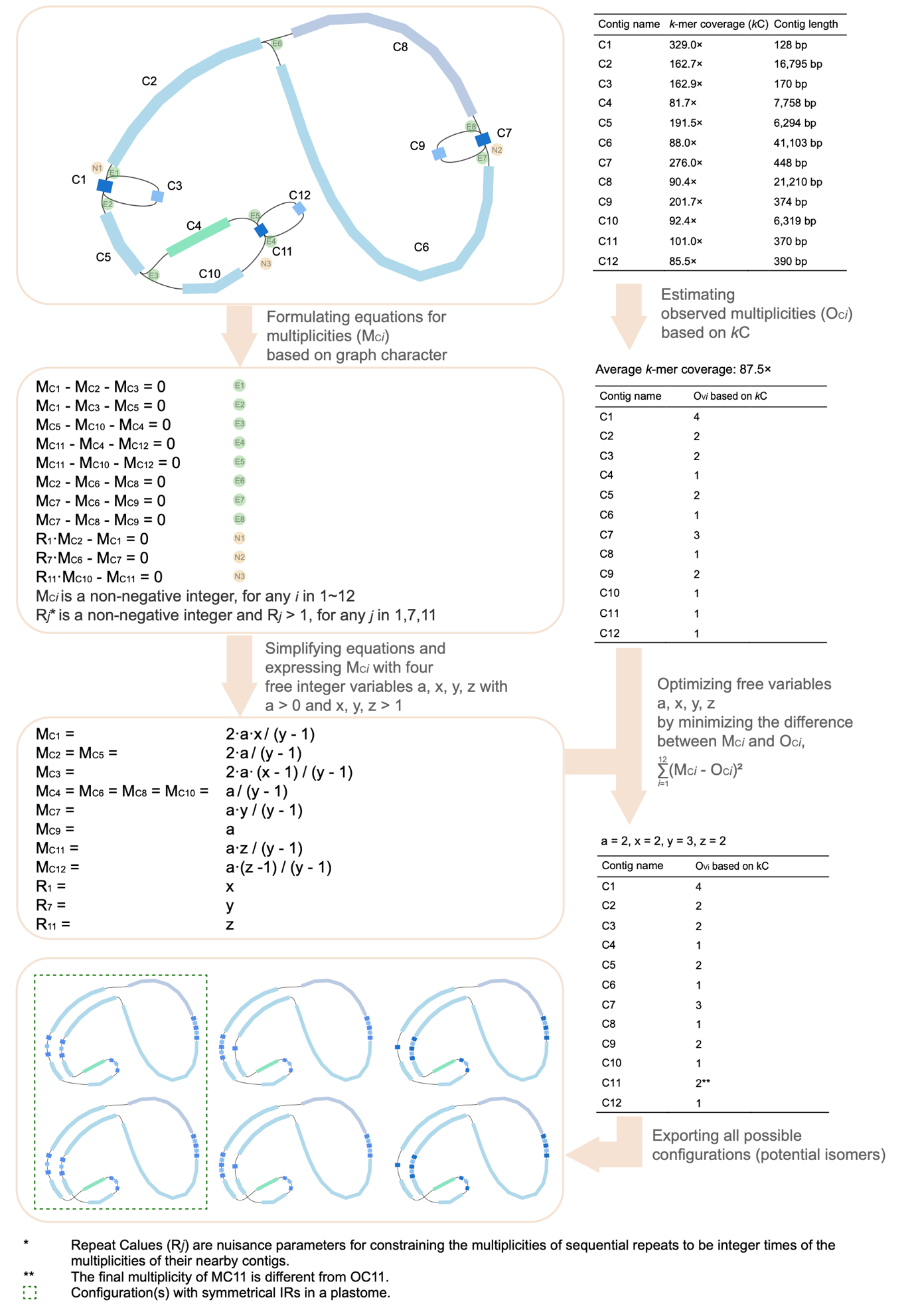
The new algorithm of contig multiplicity estimation incorporates both information of graph characteristics and contig coverage (Fig. 2).
To evaluate the accuracy of assemblies using GetOrganelle, in comparison with another popular assembler NOVOPlasty, the GetOrganelle team tested the 156 public datasets from plants, animals, and fungi. For 50 public plant WGS datasets, GetOrganelle generated a high completeness rate of 78% with default settings, significantly better than currently most popular tool NOVOPlasty which generated 16% with fine-tuning but cost slightly less computational resources.
GetOrganelle still significantly outperformed NOVOPlasty at completeness rate even when consuming comparable or less computational resources. Furthermore, NOVOPlasty generated 20%~25% wrong/false complete plastomes in K=23 and K=31 runs (Fig. 3).
In the same test, the consistency of GetOrganelle assemblies under different parameters was also better than that of NOVOPlasty assemblies.
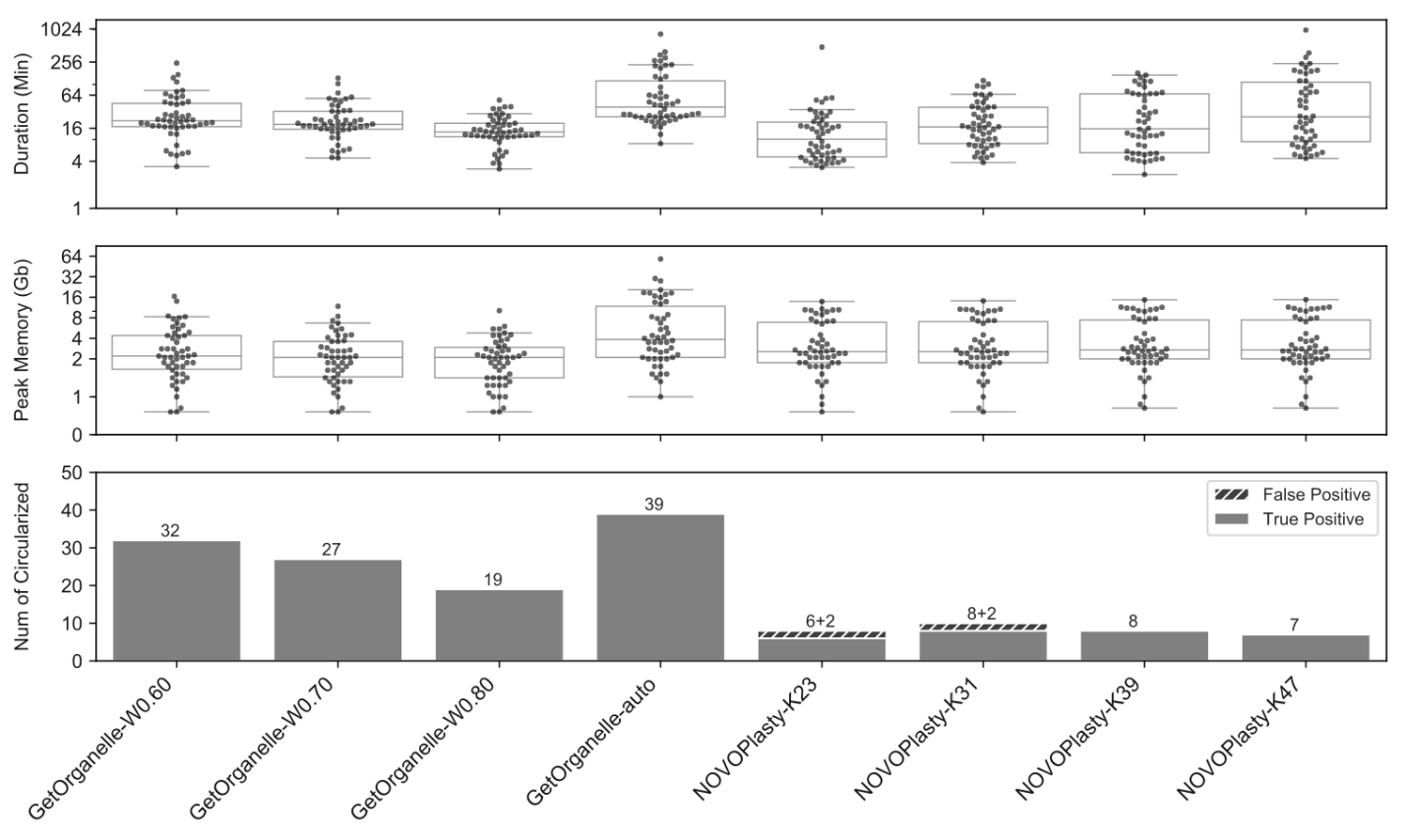
Figure 3: Comparisons of four sets of runs using GetOrganelle and four sets of runs using NOVOPlasty when assembling 50 public plant datasets. (Image by KIB)
According to the read mapping evaluation, GetOrganelle plastomes outperformed both NOVOPlasty and published plastomes from the same reads at accuracy (Fig. 4).
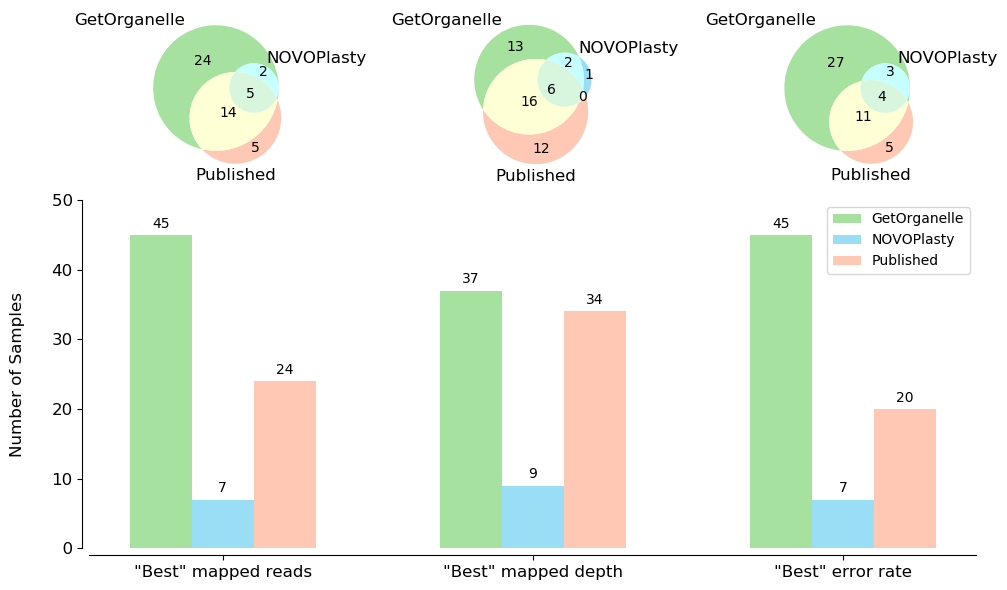
Many mistakes in the published plastomes were detected during this evaluation. For 56 animal datasets and 50 fungal datasets, GetOrganelle was generally better over NOVOPlasty in obtaining mitogenome contigs and genes.
Noteworthily, Freudenthal et al. (2020) presented a benchmark comparison of several chloroplast assembly pipelines/toolkits (including chloroExtractor, Fast-Plast, GetOrganelle, IOGA, NOVOPlasty, org.ASM) and found significant differences among those assemblers.
In their tests, GetOrganelle significantly outperformed all other assemblers in accuracy and success rate, and was recommended as the default assembler.
This study, entitled: GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes, was published online in the journal Genome Biology on September 10th, 2020.
Drs. JIN JianJun from KIB and YU Wenbin from XTBG are co-first authors, and Profs. LI Dezhu and YI Tingshuang are the corresponding authors.
This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB31000000), the National Natural Science Foundation of China [key international (regional) cooperative research project No.31720103903], the CAS Large-scale Scientific Facilities (2017-LSFGBOWS-02), the open research project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, the National Natural Science Foundation of China (31870196), and the CAS 135 Program (2017-XTBG-T03).
An early version of GetOrganelle source codewas firstly deposited in GitHub (https://www.github.com/Kinggerm/GetOrganelle) in April 2016, and the most recent update was on 27 July 2020.
Besides, the latest stable version of GetOrganelle was also deposited in Bioconda since March 2020, making the installation foolproof with conda.
Moreover, a beta version of GetOrganelle for meta-mitogenomics was recently online, and GetOrganelle for long-read sequencing is coming soon.
Noteworthily, a preprint of GetOrganelle manuscript was online available at BioRxiv in May 2018, with three revisions. To date, the preprint had more than 230 citations in GoogleScholar.
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
email: yangmei@mail.kib.ac.cn
(Editor: Yang Mei)




