Epiphytes represent a distinct group in earth carbon cycling ecosystems. Most vascular epiphytes are from the particularly species-rich orchid family (Orchidaceae), with approximately 70% of Orchidaceae species being epiphytes.
Epiphytes with crassulacean acid metabolism (CAM) photosynthesis are widespread in vascular plants, with repeated evolution of CAM photosynthesis being a key innovation for micro-ecosystem adaptation.
CAM photosynthesis is a water-conserving carbon dioxide (CO2) fixation pathway prevalently found in desert succulents and tropical epiphytes. By using a temporally separated carbon-concentrating mechanism, a CAM plant is able to photosynthesize during the day and exchange gases at night to minimize water loss.
However, the understanding of the molecular regulation of CAM photosynthesis in epiphytes remains elusive.
Researchers from the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS) have assembled a high-quality epiphytic CAM orchid genome, Cymbidium mannii, with integrating the transcriptome, proteome and metabolome analysis to reveal the multifaceted regulation mechanism in CAM epiphytes.
The C. mannii genome is 2.88 Gb in length (Figure 1) with a contig N50 of 22.7 Mb, 82.8% of which were repetitive elements. The genome size expansion in Cymbidium is mainly due to the LTR insertions, and the insertion time is consistent with the timing of the diversification of Cymbidium species (Figure 2).
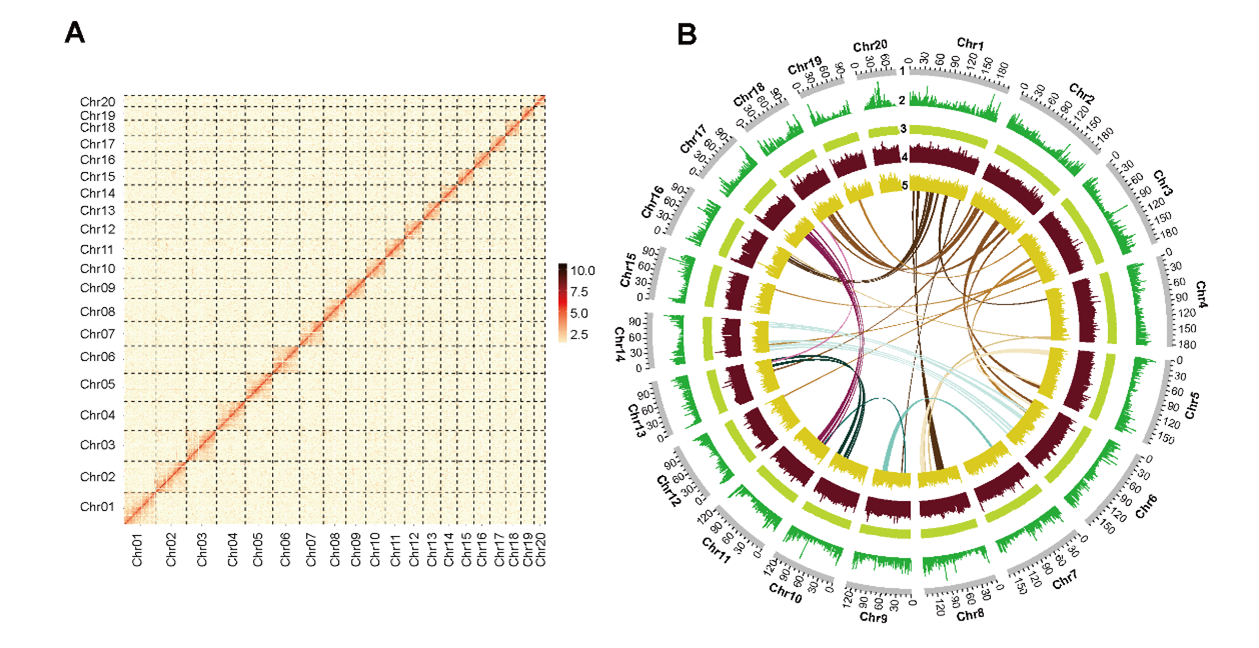
Figure 1. Chromosomal-level genomic features of C. mannii. (Image by KIB)
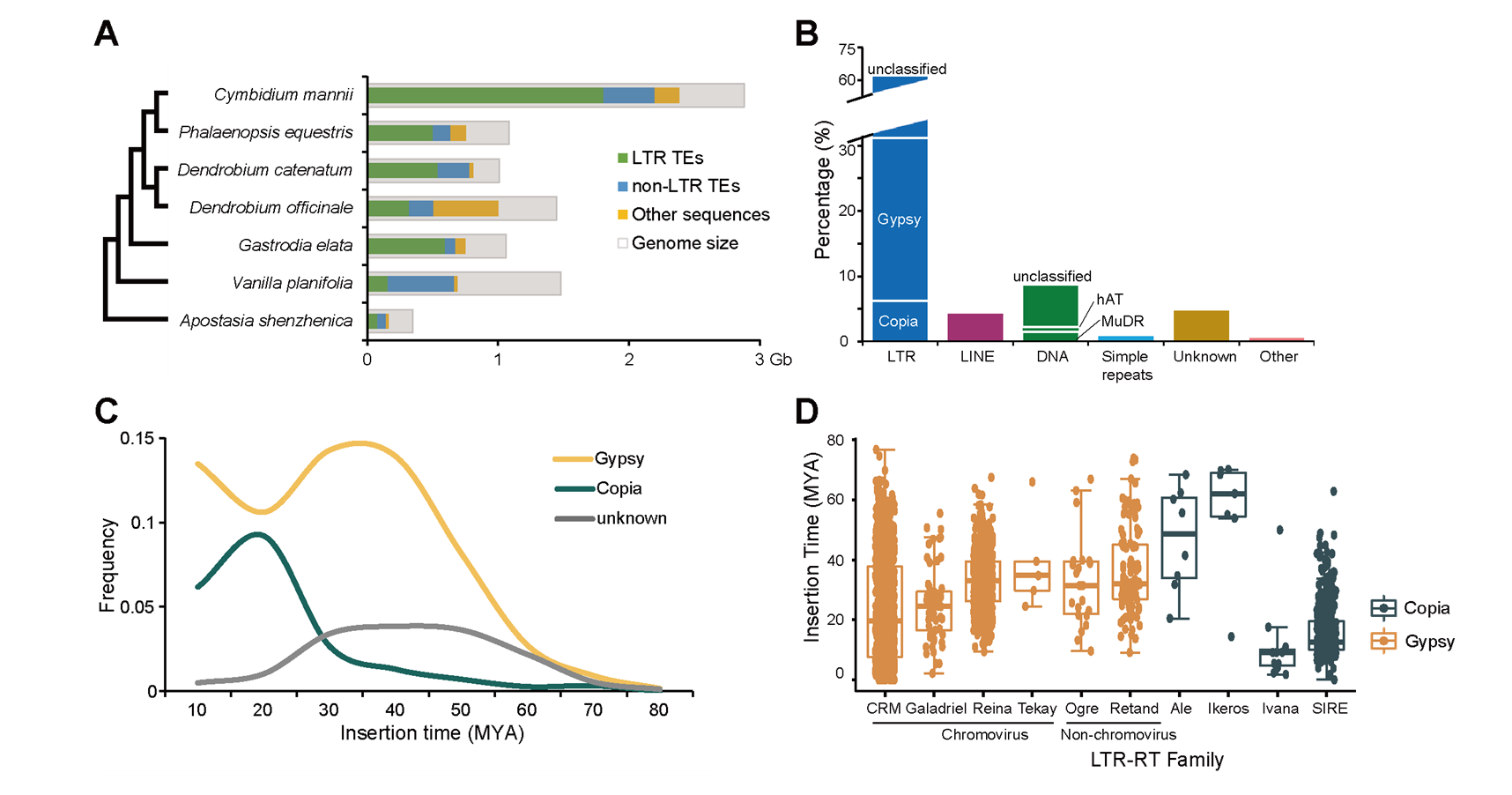
Figure 2. TE-mediated genome size expansion. (Image by KIB)
The research team found that the patterns of rhythmically oscillated metabolites, especially CAM-related products, reflect circadian rhythmicity in metabolite accumulation in epiphytes (Figure 3).
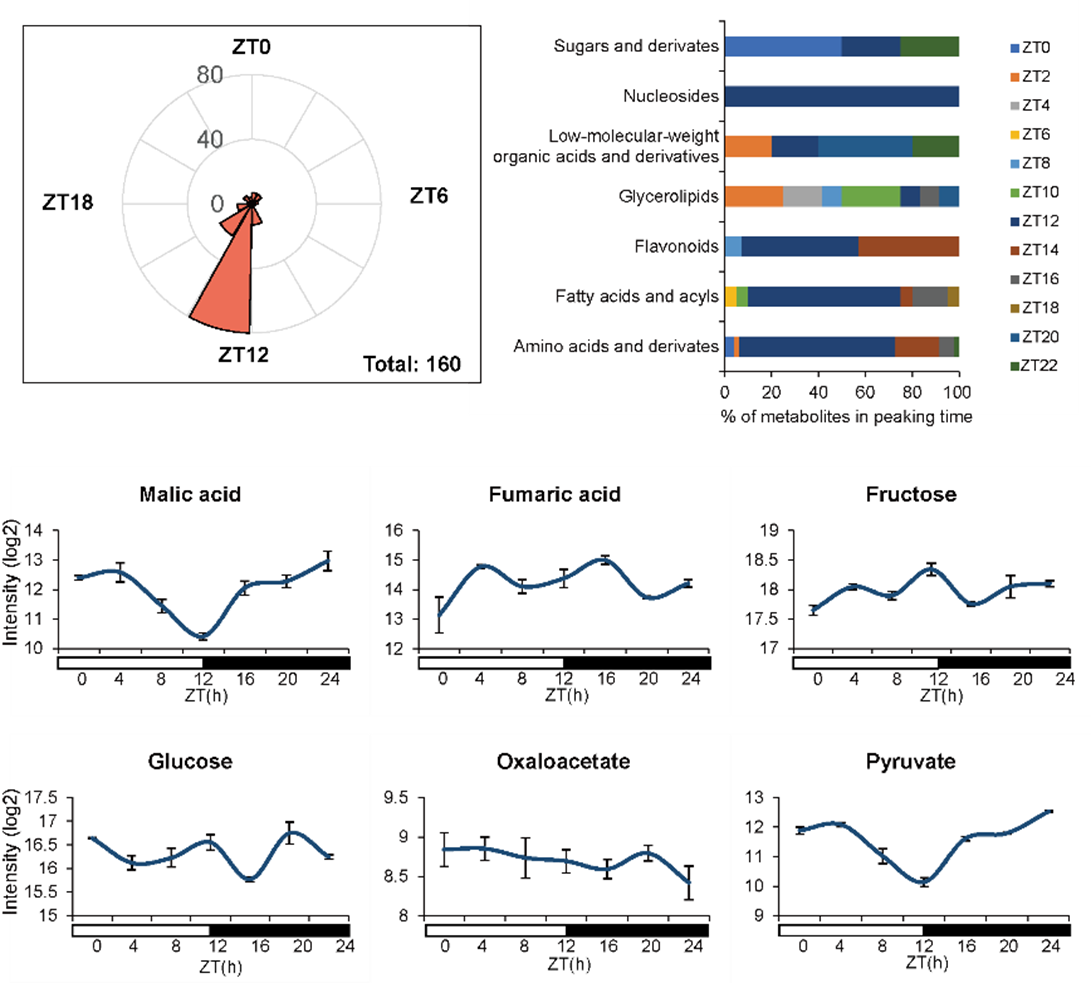
Figure 3. Rhythmic and selected CAM photosynthesis-related metabolites in C. mannii. (Image by KIB)
Genome-wide analysis of transcript and protein level regulation revealed phase shifts during the multifaceted regulation of circadian metabolism (Figure 4). The researchers show the diurnal expression of several core CAM genes (especially βCA and PPC) that might temporally fix carbon sources. The gene expression levels of NADP-ME and PPDK in C. mannii were highly concordant with those in K. fedtschenkoi and S. album, revealing that they all preferred to utilize NADP-ME and PPDK pathways for decarboxylation (Figure 5).
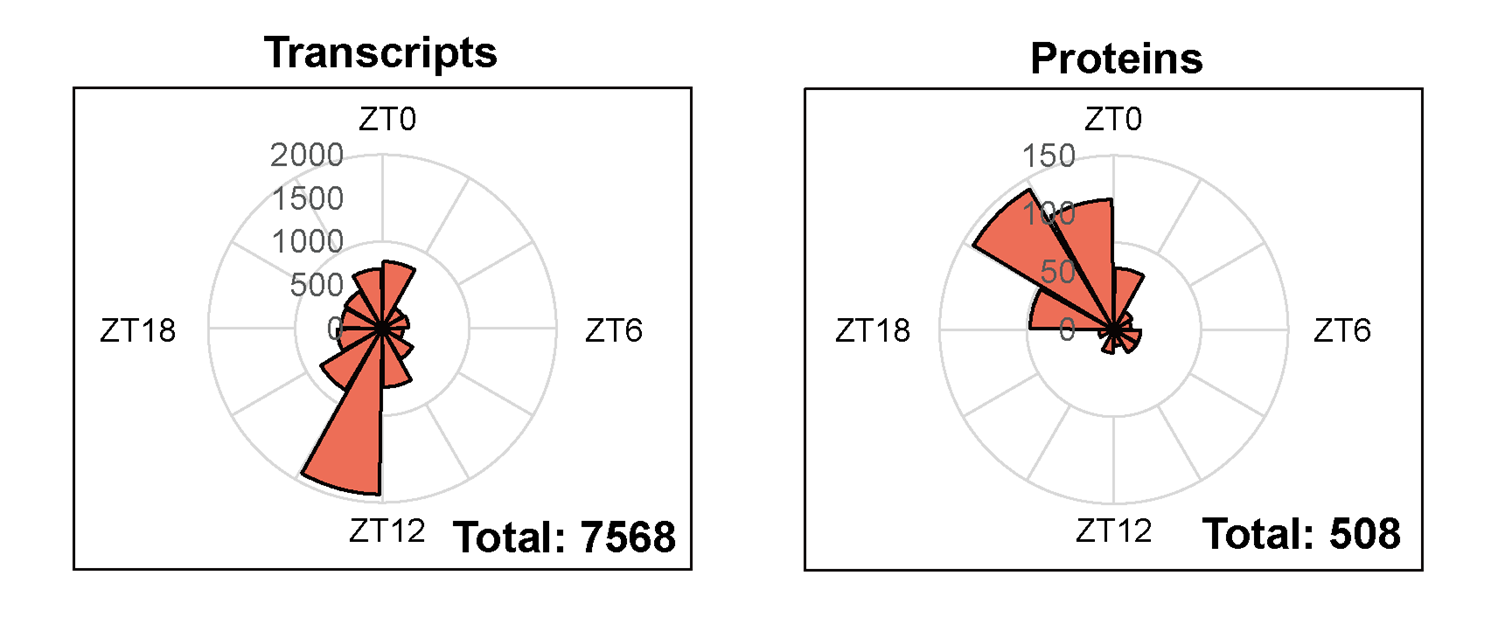
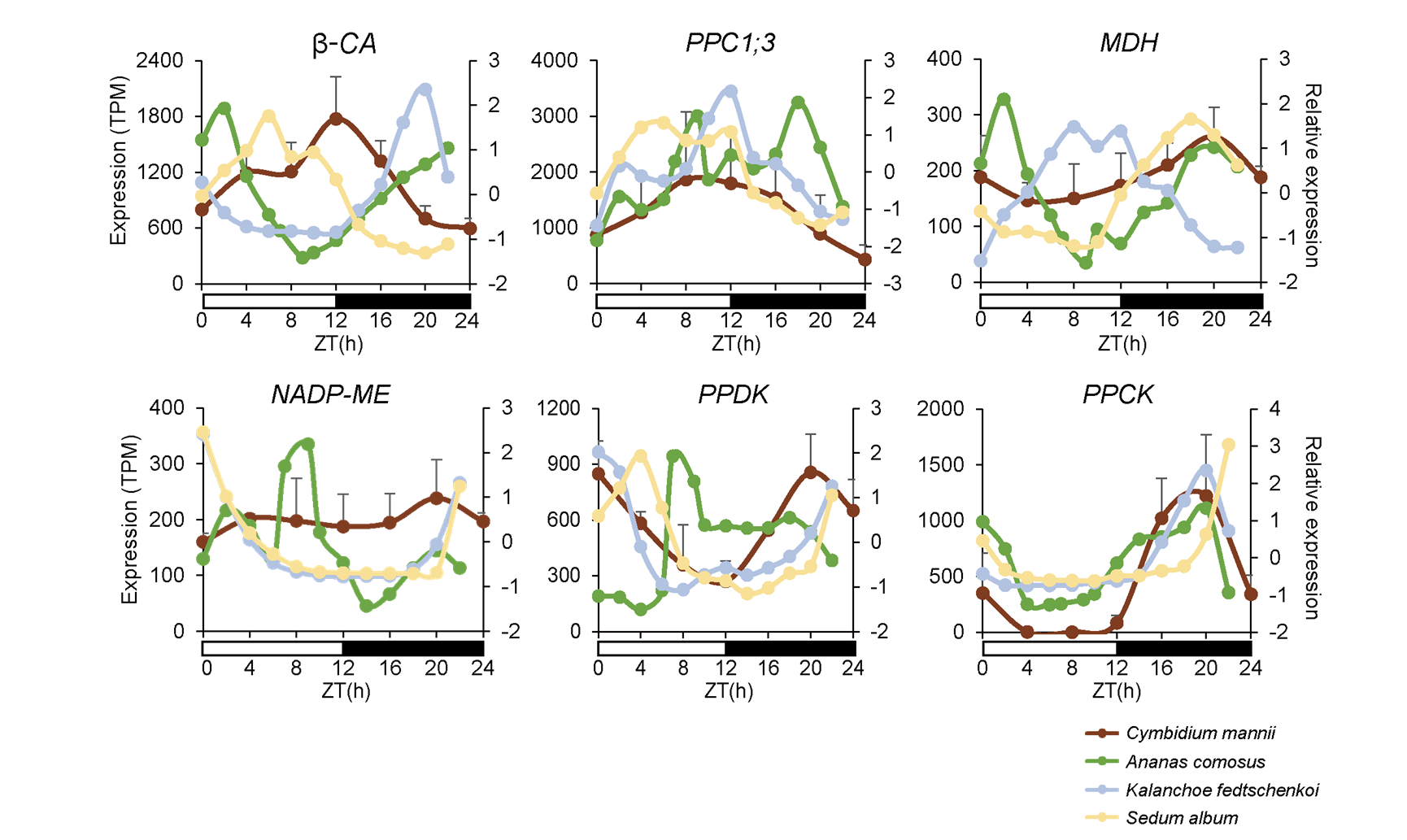
Figure 5. Selected cycling core CAM gene expression in C. mannii and compared with terrestrial CAM plants. (Image by KIB)
Since the circadian clock-associated genes and CREs play a crucial role in regulating the CAM pathway, researchers explored the circadian clock and CAM gene expression pattern during the diel cycle and found the light-responsive CREs in promoter regions possibly involving in photoperiodism (Figure 6).
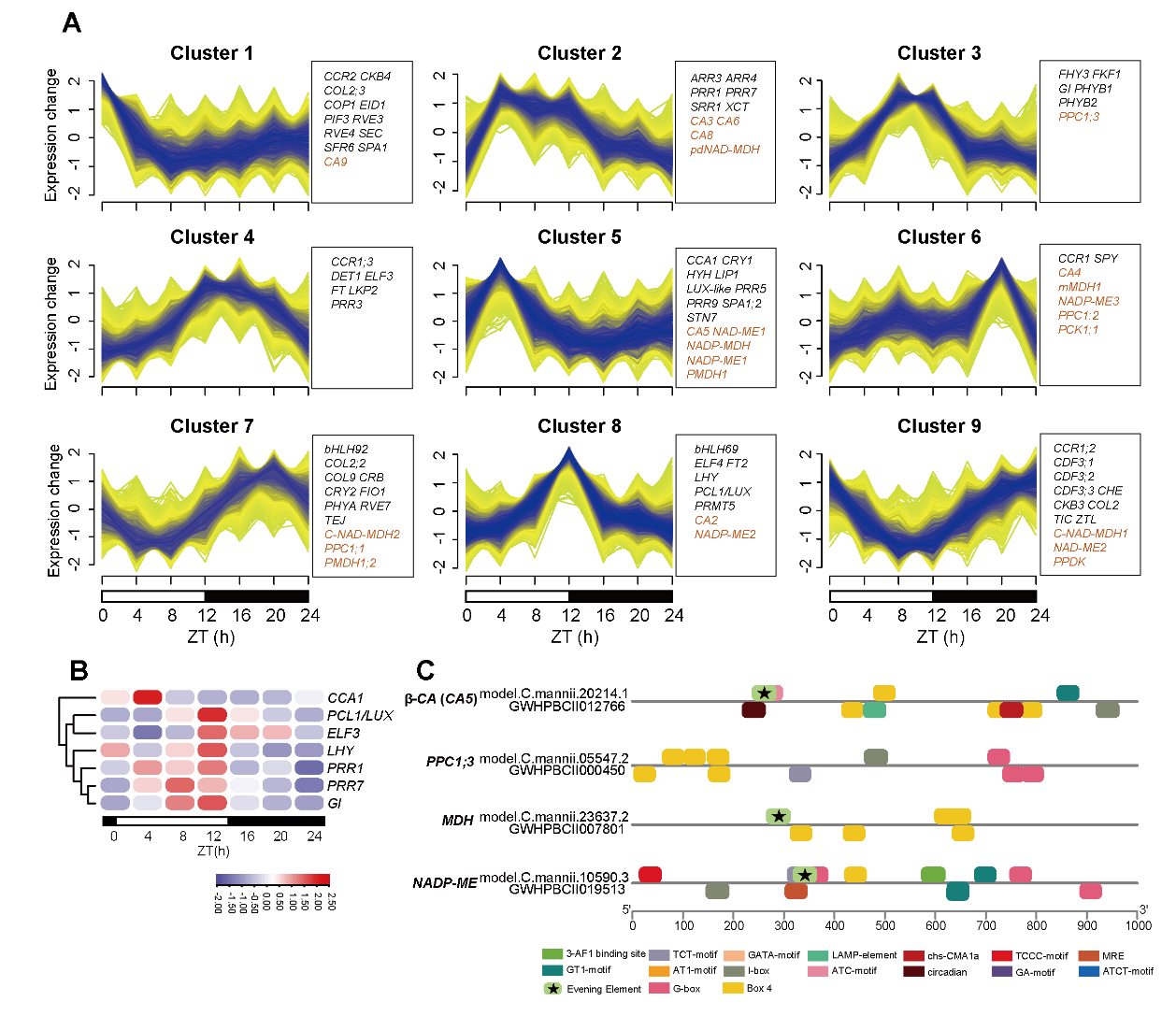
Figure 6. Core circadian clock gene expression level and CREs in selected CAM genes. (Image by KIB)
This research was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, the Project for Innovation Team of Yunnan Province, and the Science and Technology Basic Resources Investigation Program of China.
Research Results have been published on Plant Communications entitled "High-quality Cymbidium mannii genome and multifaceted regulation of crassulacean acid metabolism in epiphytes".
(Editor:YANG Mei)




