As a main driving force in evolution, polyploidy (genome duplication) is ubiquitous across various evolutionary stages of the flowering plant tree of life. However, the interactions between the ancestral genomes in a polyploid nucleus, frequently involving subgenome dominance, are poorly understood.
Bamboos exhibit great diversity in species and morphology, consisting of a minor herbaceous, mostly diploid clade (ca. 126 species) and three major polyploid clades of woody plants (ca. 1,576 species). As one of the most important lineages of plants that has adapted to a forest habitat, woody bamboos (WBs) exhibit distinctive biological traits, such as rapid growth (up to 114.5 cm daily) and synchronous, usually monocarpic, flowering (~30–60 years).
The research team led by Prof. LI Dezhu from the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS), in collaboration with scientists from the United States, proposed a refined model for the origins and polyploidizations of woody bamboos.
Their studies revealed recurrent hybridization events between diploid ancestors of woody lineages followed by polyploidization, together with introgression between ancestral woody and herbaceous lineages, early in the evolution of bamboos (Fig. 1).
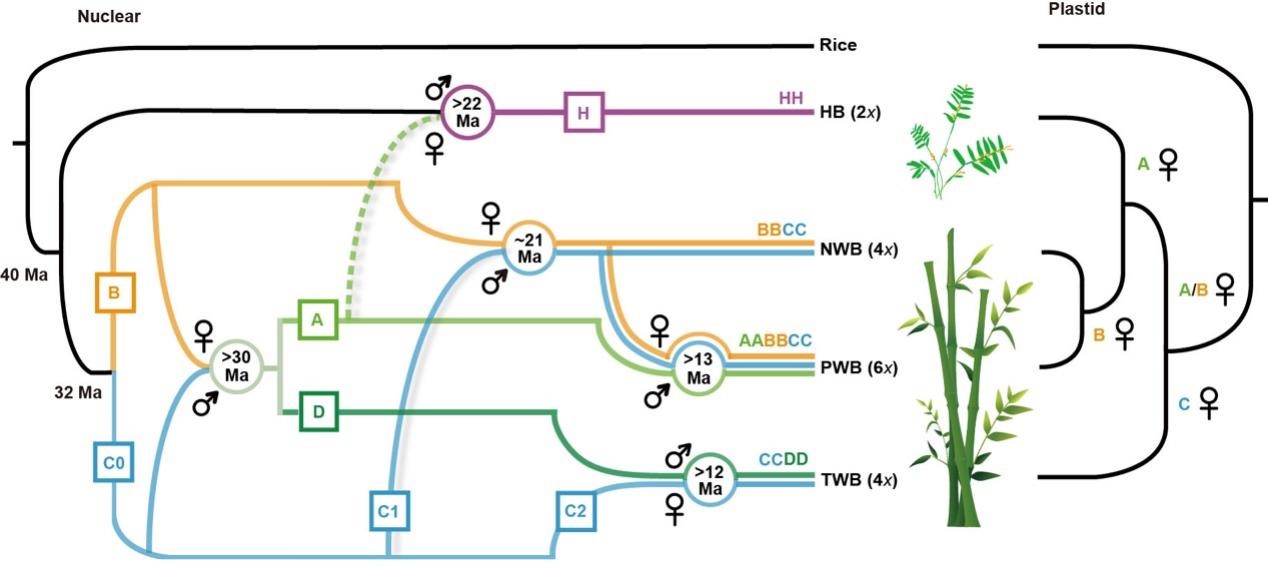
Figure 1. Model for the origins and evolutionary history of diploid bamboo ancestors and the polyploidization events in three woody clades. (Image by KIB)
LI’s team has long been dedicated to systematic and evolutionary studies of bamboos. In their previous work reporting draft genomes for four bamboo species, they proposed a reticulate origin for WBs involving hybridization and polyploidization.
“To conduct a more comprehensive study of bamboos across different ploidy levels and broad phylogenetic diversity, we sampled two herbaceous bamboos and three representative species from each woody lineage,” said LI.
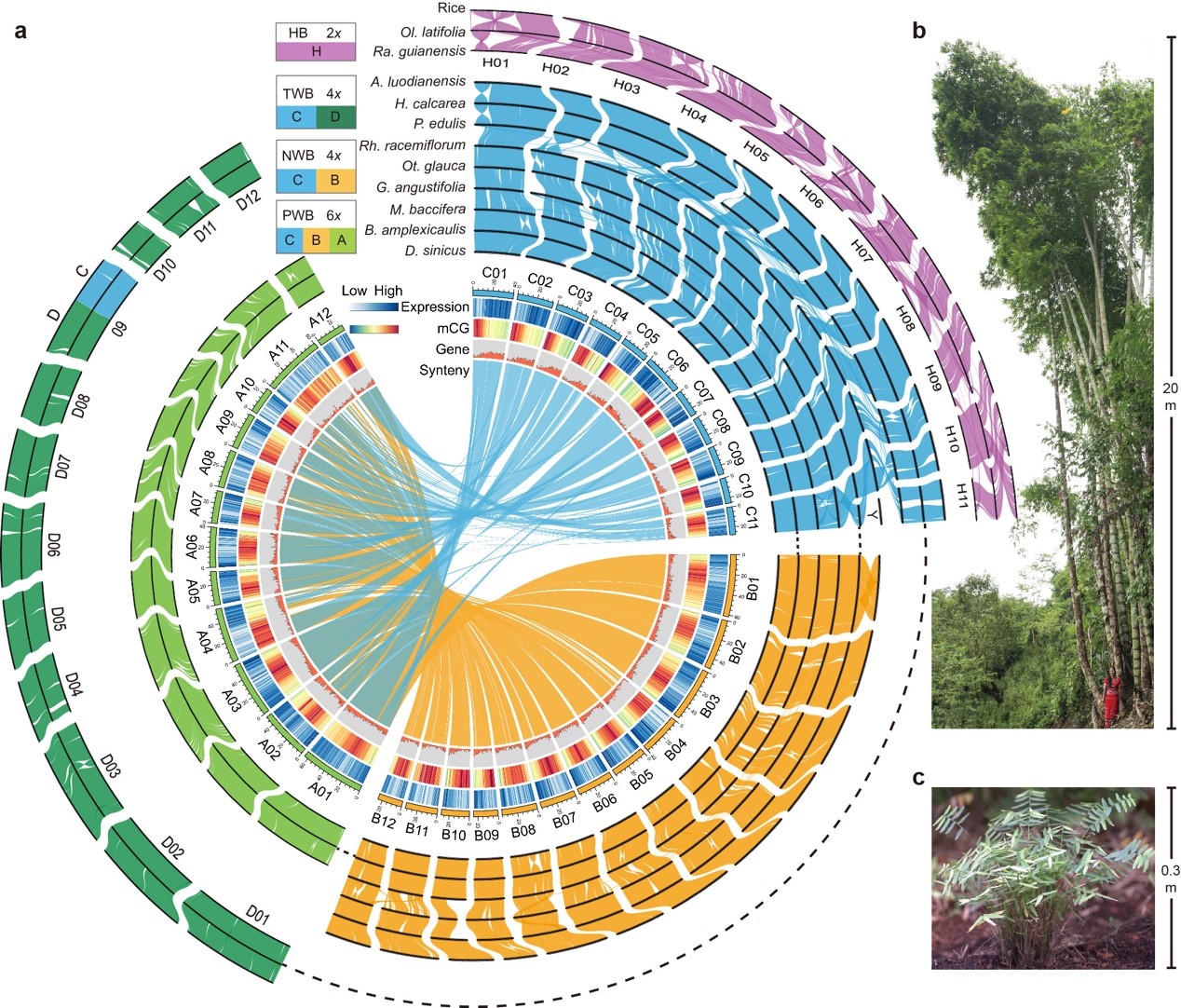
Figure 2. Overview of syntenic landscape of sequenced genomes of two herbaceous bamboos and three representative species from each woody clade, and size difference between Dendrocalamus sinicus (b), a hexaploid woody species, and Raddia guianensis (c), a diploid herbaceous species. (Image by KIB)
Further research found that the WBs exhibit stunning karyotype stability by maintaining the ancestral state of 12 chromosomes of the grass family at the subgenome level, despite 12 to 20 Ma since polyploidization and subsequent large-scale species diversification.
"Our study suggested unambiguous dominance of the C subgenomes in two tetraploid lineages, as reflected in a series of features including genome size, gene fractionation, genomic rearrangements, transposable elements and gene expression,” said LI.
However, the pattern of dominance in the hexaploid lineage is more dynamic (Figure 3), with a gradual shift of dominance from the C to the A subgenome.
Further analyses revealed that the dominant C subgenome, together with the A subgenome in the hexaploid clade, contributed the most to the evolution of distinctive traits in WBs and possibly their adaptive radiation into a forest habitat.
This work highlights the utility of using clade-wide genome sequence assemblies to advance our understanding of subgenome evolution in polyploid plants.
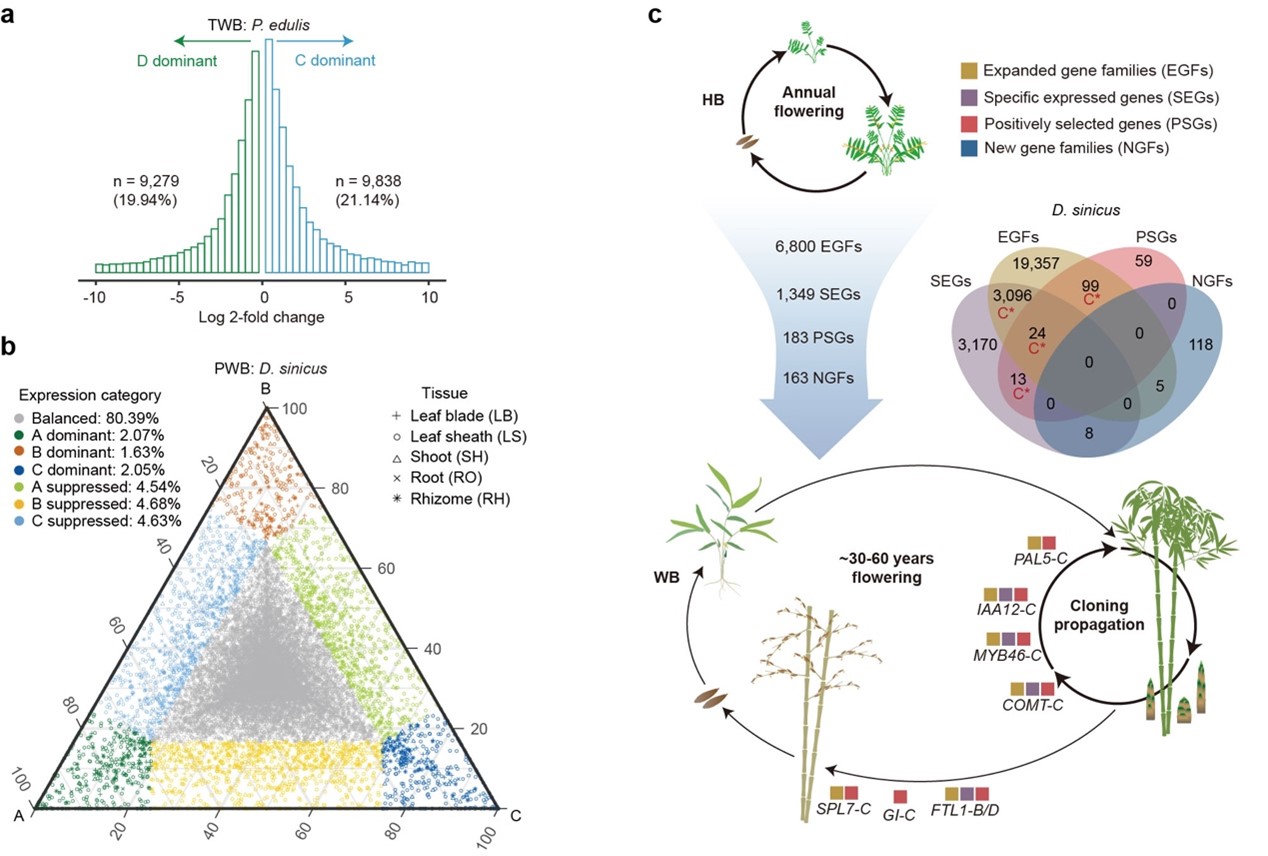
Figure 3. Histograms (a) and ternary plot (b) of homoeologs for expression bias in representative species of tetraploid and hexaploid clades of woody bamboos and life history transition from herbaceous to woody bamboos with underlying genetic alterations (c). (Image by KIB)
Results of this work were published online in Nature Genetics, entitled "Genome assemblies of 11 bamboo species highlight diversification induced by dynamic subgenome dominance".
Drs. MA Pengfei, LIU Yunlong, GUO Cen (currently working at the CAS’s Xishuangbanna Tropical Botanical Garden), JIN Guihua, and GUO Zhenhua from KIB are the co-first authors of the paper, with Prof. LI Dezhu as the corresponding author. Prof. Lynn G. Clark from the Iowa State University, Dr. Elizabeth A. Kellogg from the Danforth Plant Science Center, Prof. Heffrey L. Bennetzen from the University of Georgia, and Profs. Pamela S. Soltis & Douglas E. Soltis from the University of Florida have also engaged in this work.
This study was supported by the CAS’s Strategic Priority Research Program,the National Natural Science Foundation of China, and the Leading Talents Program of Yunnan Province, China.
(Editor:YANG Mei)




