Genomic variation, environmental adaptation and feralization in ramie, an ancient fiber crop were revealed by Prof. LI Dezhu’s team at the Kunming Institute of Botany of the Chinese Academy of Sciences (KIB/CAS) in collaboration with researchers from the Xishuangbanna Tropical Botanical Garden,CAS, Institute of Bast Fiber Crops of the Chinese Academy of Agricultural Sciences, University of Southampton, University of Edinburgh, Royal Botanic Gardens Kew, and University of Toronto.
The article was online published in Plant Communications, a Cell Press partner journal in plant science.
Feralization is the evolutionary process by which domesticated crops or livestock re-acquire some wild-like traits and escape from intensive management to form independent reproducing populations. Feralization has interested biologists since Darwin, not only because of the implications for evolution but also because feral populations can become invasive and have severe ecological or agricultural impacts.
On the other hand, feral populations might be significant reservoirs of genetic diversity for crop breeding. A greater understanding of feral populations at the genetic level might therefore help to both mitigate their impacts as weeds or pests and evaluate them as potential genetic reservoirs.
However, despite increasing attention, the evolutionary mechanisms underlying feralization remains poorly understood. At least 14 feralization events in crops have been suggested, but only 4 crops have been investigated at the genomic level.
The basis of adaptation and ecological niche range in plants escaping cultivation have yet to be investigated.
To address this, the research team produced a chromosome-scale de novo genome assembly of feral ramie and investigated structural variations between feral and domesticated ramie genomes.
915 accessions from 23 countries were gathered, comprising cultivars, major landraces, native wild accessions, and feral populations. Based on whole genome resequencing of these accessions, the most comprehensive ramie genomic variation map to date was constructed.
Phylogenetic, demographic, and admixture signal detection analyses indicate that feral ramie is of exoferal or exo-endo origin, i.e., descended from hybridization between domesticated ramie and wild progenitor or ancient landrace .
Feral ramie has greater genetic diversity than wild progenitor or domesticated ramie, and genomic regions affected by natural selection during feralization are different from those under selection during domestication.
Ecological analyses showed that feral and domesticated ramie have similar ecological niches which are substantially different from the niche of the wild progenitor, and three environmental variables were associated with habitat-specific adaptation in feral ramie.
These findings advance the understanding of feralization, providing a scientific basis for the excavation of new crop germplasm resources and offering novel insights into the evolution of feralization in nature.
This work was supported by the CAS Strategic Priority Research Program of the National Natural Science Foundation of Chinaand the CAS Youth Innovation Promotion Association.

Fig. 1 Morphological comparison among three varieties of ramie. A-C, Boehmeria nivea var. nivea. D-F, B. nivea var. strigosa; D, habit with branched stem; E, green abaxial leaf blade; F, patent strigose stem and partly connate stipule. G-I, B. nivea var. tenacissima. (Image by KIB)
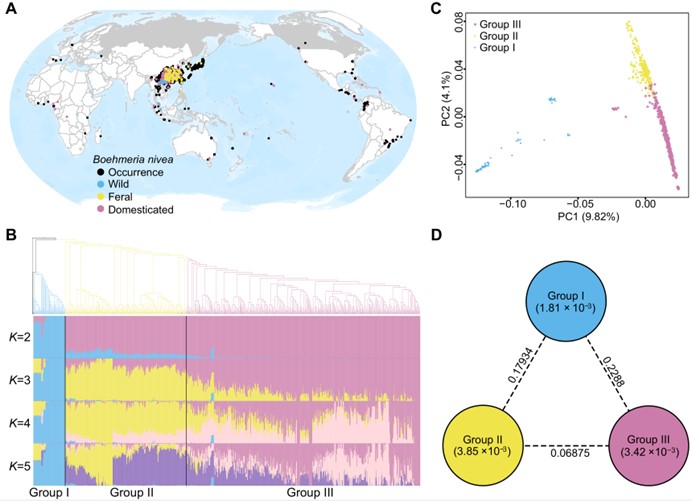
Fig. 2. Population structure and genetic diversity of ramie. (Image by KIB)
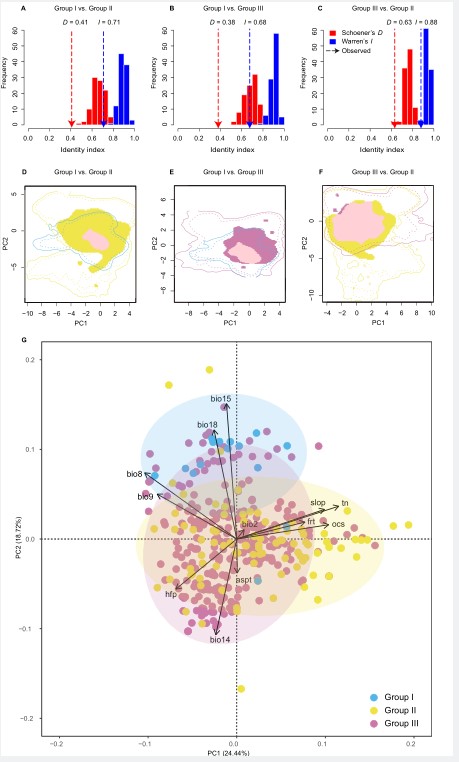
Fig. 3. Ecological analyses results. (Image by KIB)
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
email: yangmei@mail.kib.ac.cn
(Editor: Yang Mei)




