Modern cultivated strawberry (Fragaria × ananassa) originates from interspecific hybridization between two octoploid wild species—F. virginiana and F. chiloensis. However, the divergence in centromere characteristics and evolutionary trajectories between the wild and cultivated octoploids has remained poorly understood.
In a recent study published in Genome Biology, a research team led by ZHU Andan from Kunming Institute of Botany, Chinese Academy of Sciences (KIB/CAS), elucidated centromere divergence and evolutionary patterns in octoploid strawberries. The team achieved this breakthrough by generating a strawberry CENH3-specific antibody and employing advanced genome assembly techniques.
The researchers evaluated different genome phasing strategies and determined that the Trio-binning approach facilitated global phasing with moderate contiguity, while the Hi-C-based method yielded local phasing with superior contiguity. Leveraging these insights, they integrated the strengths of both methods to develop a novel “two-step” phasing assembly pipeline. This innovation enabled the construction of a nearly complete, globally phased genome for the octoploid strawberry cultivar “EA78”.
Notably, the high-quality genome assembly of “EA78” demonstrated centromere turnover, with six chromosomes exhibiting loss of the canonical 147-bp centromeric satellite sequence—a finding corroborated by fluorescence in situ hybridization (FISH) assays. To further explore this phenomenon, the team utilized their CENH3-specific antibody to identify six neocentromeres and three new types of centromeric satellites in “EA78”. Sequence similarity density analysis between canonical centromeres and neocentromeres demonstrated a bimodal distribution pattern, indicating substantial genetic divergence between these two centromere types.
Intriguingly, comparative analyses revealed that centromere length and satellite abundance exhibited significant expansion trends in both wild and cultivated octoploids relative to their diploid strawberry ancestor, F. vesca. This suggests that hybridization events and domestication processes may collectively drive centromere expansion.
This study provides critical insights into the rapid evolution of centromeres in octoploid strawberries, advancing our understanding of genomic adaptation in polyploid plants and highlighting the dynamic interplay between hybridization, domestication, and centromere plasticity.
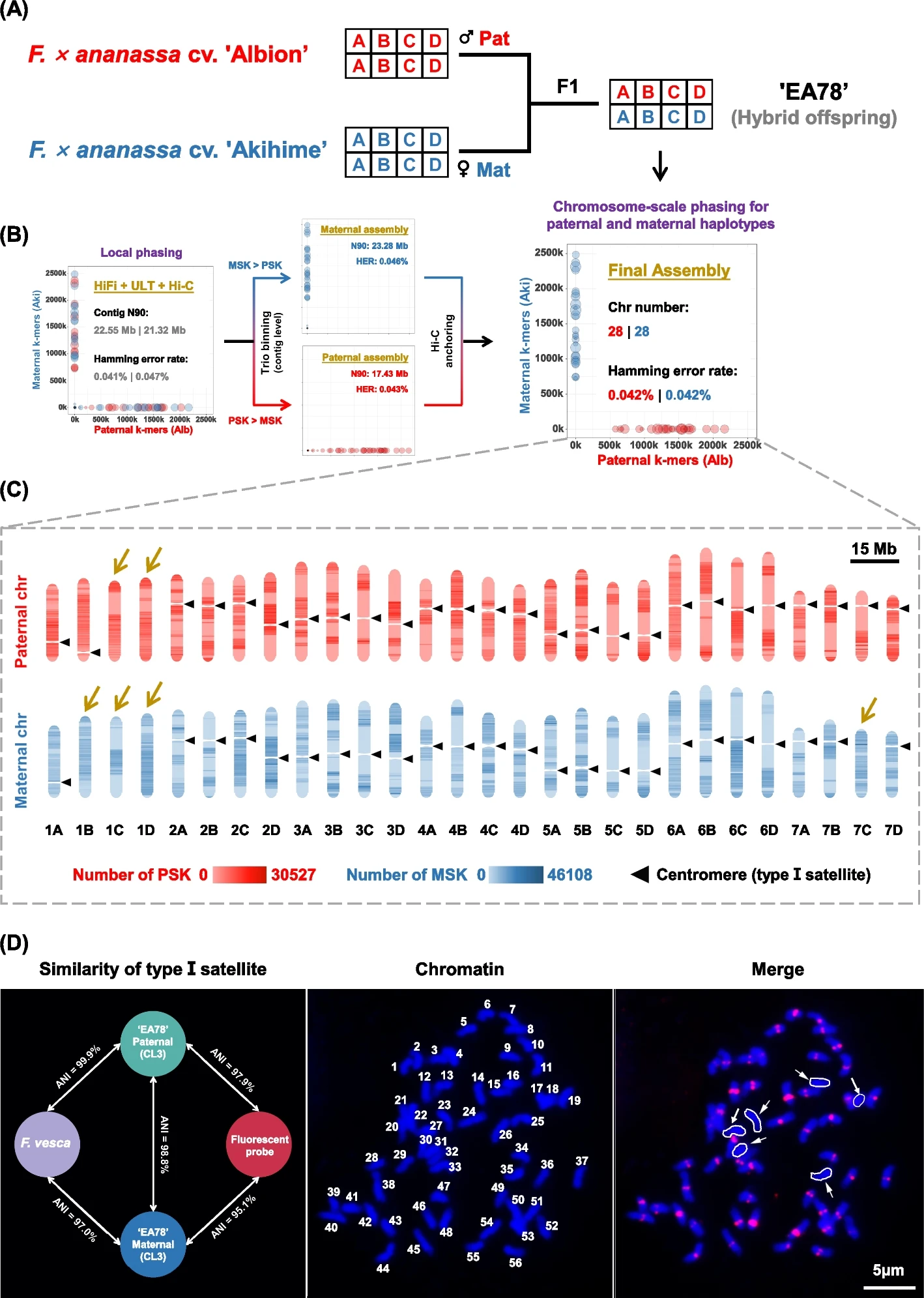
The octoploid “EA78” genome assembly and the turnover of centromeric satellites. A Plant materials and experimental design. The parental cultivars are both octoploids with four subgenomes (A, B, C, D), following the subgenome designation of Jin et al. (2023) [14]. Pat: paternal, Mat: maternal. B A two-step phased assembly strategy for achieving global phasing by integrating HiFi, Hi-C, ONT Ultra-long sequencing data, and pedigree information. Note: UL: ultra-long. PSK: paternal-specific k-mers. MSK: maternal-specific k-mers. HER: hamming error rate. C Construction of a nearly complete and fully phased assembly of the “EA78” genome. The genomic locations of putative centromeres were shown in each chromosome. Arrows highlighting six chromosomes lacking canonical centromeric repeats. D The characterization of canonical centromeric repeats. Left: sequence similarity (based on average nucleotide identity, ANI) among centromere repeats of F. vesca, paternal and maternal assemblies, and sequence for fluorescent prove design. Middle: “EA78” chromatin number statistics. Right: FISH assay using the type I centromeric satellites as probes (scale bar = 5 μm) (Image by KIB)
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
email: yangmei@mail.kib.ac.cn
Editor:YANG Mei




