Urticaceae is a large family within the order Rosales of angiosperms, currently comprising 54 genera with approximately 2,600 species. The family is distributed worldwide, with its species diversity centered in tropical regions, particularly tropical Asia and Central and South America, while temperate regions exhibit relatively lower species diversity. Morphologically, Urticaceae exhibits high diversity in life forms, inflorescence types, and stigmatic structures of female flowers.
Many species hold significant economic value, with some being important fiber crops and others showing potential pharmacological applications. Throughout its taxonomic history, systematic studies based on morphology have been controversial due to the presence of stinging hairs in many species and their typically minute, structurally complex flowers. Currently, the family is widely accepted as comprising six tribes, a classification supported by molecular phylogenetic studies (Fig. 1). However, molecular phylogenetic research at the familial level remains limited. Previous studies were often constrained by the use of few molecular markers and limited taxon sampling, failing to fully resolve the internal phylogenetic relationships of the family and thus hindering in-depth exploration of its diversity and spatiotemporal evolutionary history.
To establish a robust phylogenetic framework for Urticaceae, a collaborative effort led by Prof. LI Dezhu’s team—alongside Prof. YI Tingshuang’s group at the Chinese Academy of Sciences (CAS), the Royal Botanic Gardens, Kew, the University of Edinburgh, the Smithsonian Institution’s National Museum of Natural History, and other institutions—has recently published a paper in Plant Diversity (https://doi.org/10.1016/j.pld.2025.12.003).
This work presents the most comprehensive phylogenomic framework and taxonomic revision for Urticaceae to date. Through global sampling and integration of data from both literature and public databases such as NCBI, the study assembled chloroplast genome and nuclear ribosomal DNA (nrDNA) sequences from 485 samples, representing 345 species across all 54 genera within the family. Phylogenomic analyses were performed using 87 protein-coding genes from chloroplast genomes and nrDNA data.
The study further revised the family's classification by incorporating published phylogenetic relationships from the angiosperm "353 nuclear genes" dataset.The results showed that chloroplast genome data effectively resolved phylogenetic relationships at the generic level and above within Urticaceae, with the monophyly of the four major clades and six tribes receiving strong support. Compared to the phylogenetic tree based on the nrDNA dataset, the two were largely consistent regarding the monophyly of genera and relationships above the generic level, with the main differences being more nodes with lower support values in the nrDNA tree (Fig. 2).
Comparing the phylogenetic relationships based on chloroplast genomes from this study with those from Baker et al. (2022) based on the angiosperm 353 nuclear genes dataset (which included 74 samples covering 73 species from 52 genera), the study found high consistency in intergeneric relationships (Fig. 3). Building on this, and incorporating morphological characteristics and relevant literature, the researchers conducted a detailed taxonomic revision of the six existing tribes in Urticaceae and proposed a seventh tribe—Sarcochlamydeae.
This tribe belongs to Clade IV and is sister to the tribe Cecropieae. Furthermore, the study identified a new genus, Chiajuia, belonging to the tribe Cecropieae. Species within this genus were previously identified as Debregeasia wallichiana Wedd. The genus is named in honor of the renowned Chinese plant taxonomist Chen Jiarui, acknowledging his significant contributions to taxonomy of Urticaceae (Fig. 4). Overall, this study provides a comprehensive phylogenetic analysis of Urticaceae based on phylogenomic data, laying a foundation for further research into areas such as fiber crop domestication, biogeography, and diversification history within the family.
This work was supported by the National Natural Science Foundation of China (42171071), Yunnan Fundamental Research Projects (202401AT070190), the Top-notch Young Talents Project of Yunnan Provincial “Ten Thousand Talents Program” (YNWR-QNBJ-2020-293), CAS “Light of West China” Program, Key Research Program of Frontier Sciences, CAS (ZDBS-LY-7001), the Yunnan Revitalization Talent Support Program: Yunling Scholar Project (XDYC-YLXZ-2024-0021), the Science and Technology Basic Resources Investigation Program of China (No. 2019FY100900), the National Natural Science Foundation of China, key international (regional) cooperative research project (No. 31720103903), and the China Scholarship Council (202304910135 and 202304910138).
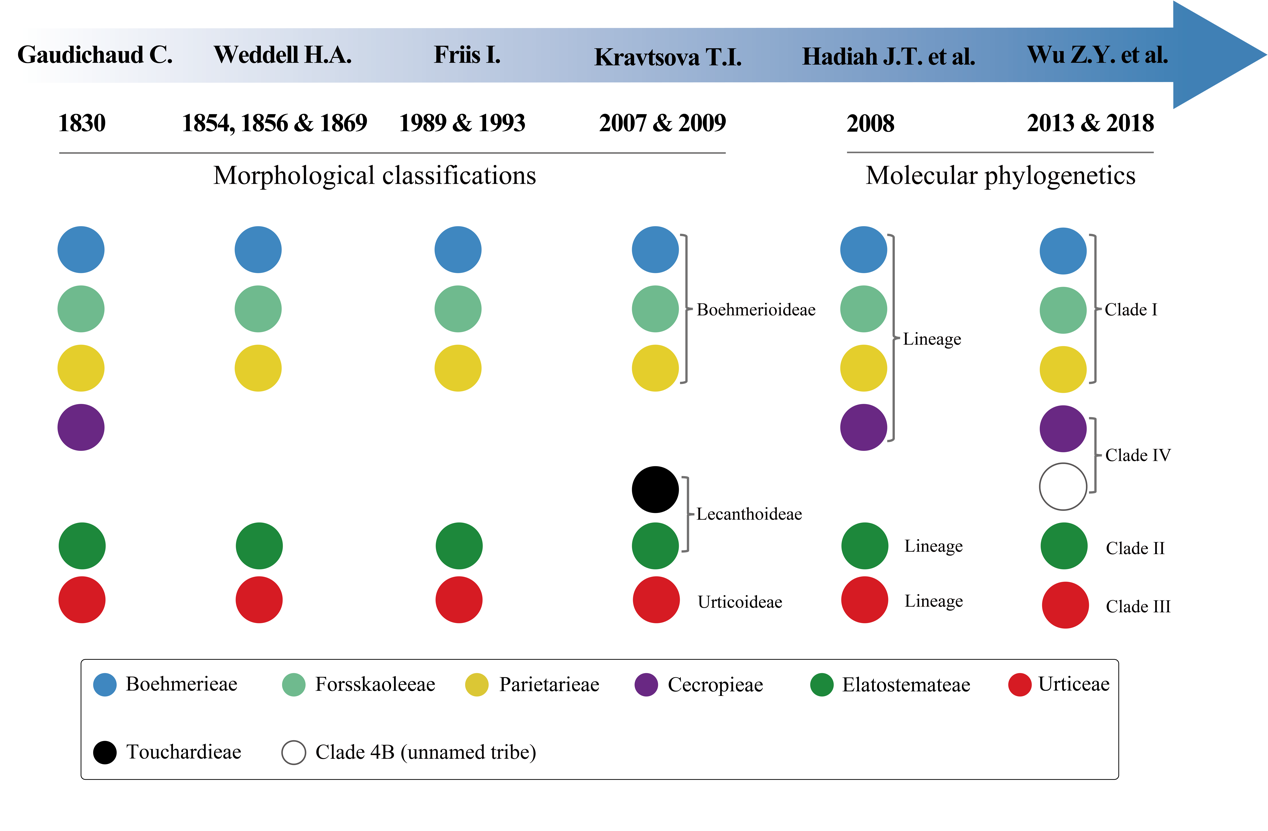
Figure 1. Historical development of major classification systems for Urticaceae, covering the progression from early morphological taxonomy to recent molecular phylogenetic studies (Image by KIB)
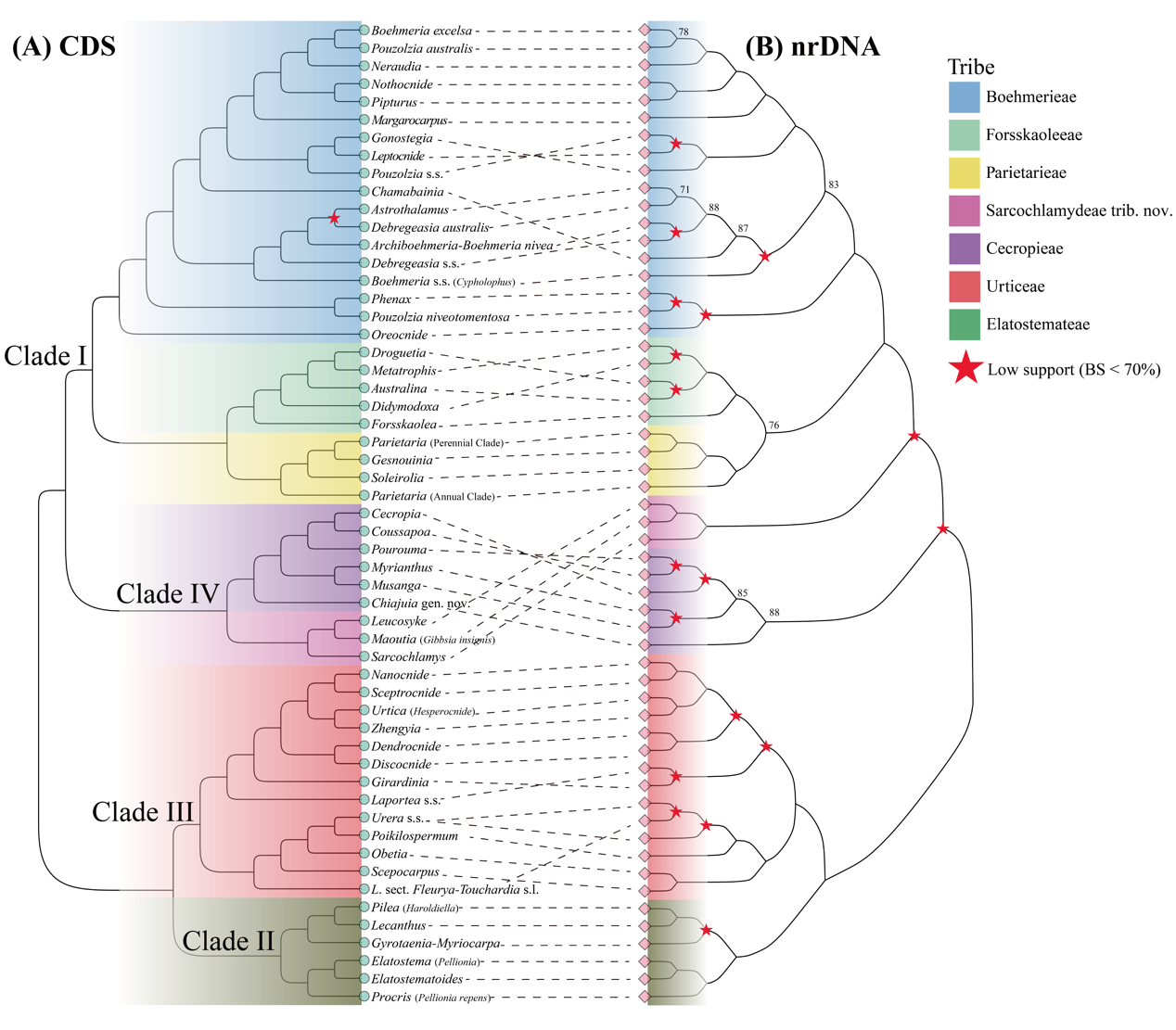
Figure 2. Comparison of Urticaceae phylogenetic trees constructed using Maximum Likelihood based on concatenated data from 87 chloroplast protein-coding genes (CDS) (A) and nuclear ribosomal DNA (nrDNA) (B). Black dashed lines connect genera or clades consistent in both topologies. All nodes have bootstrap support (BS) > 90% unless specified; only nodes with BS ≤ 90% are annotated, and nodes with BS < 70% are marked with red stars. (Image by KIB)
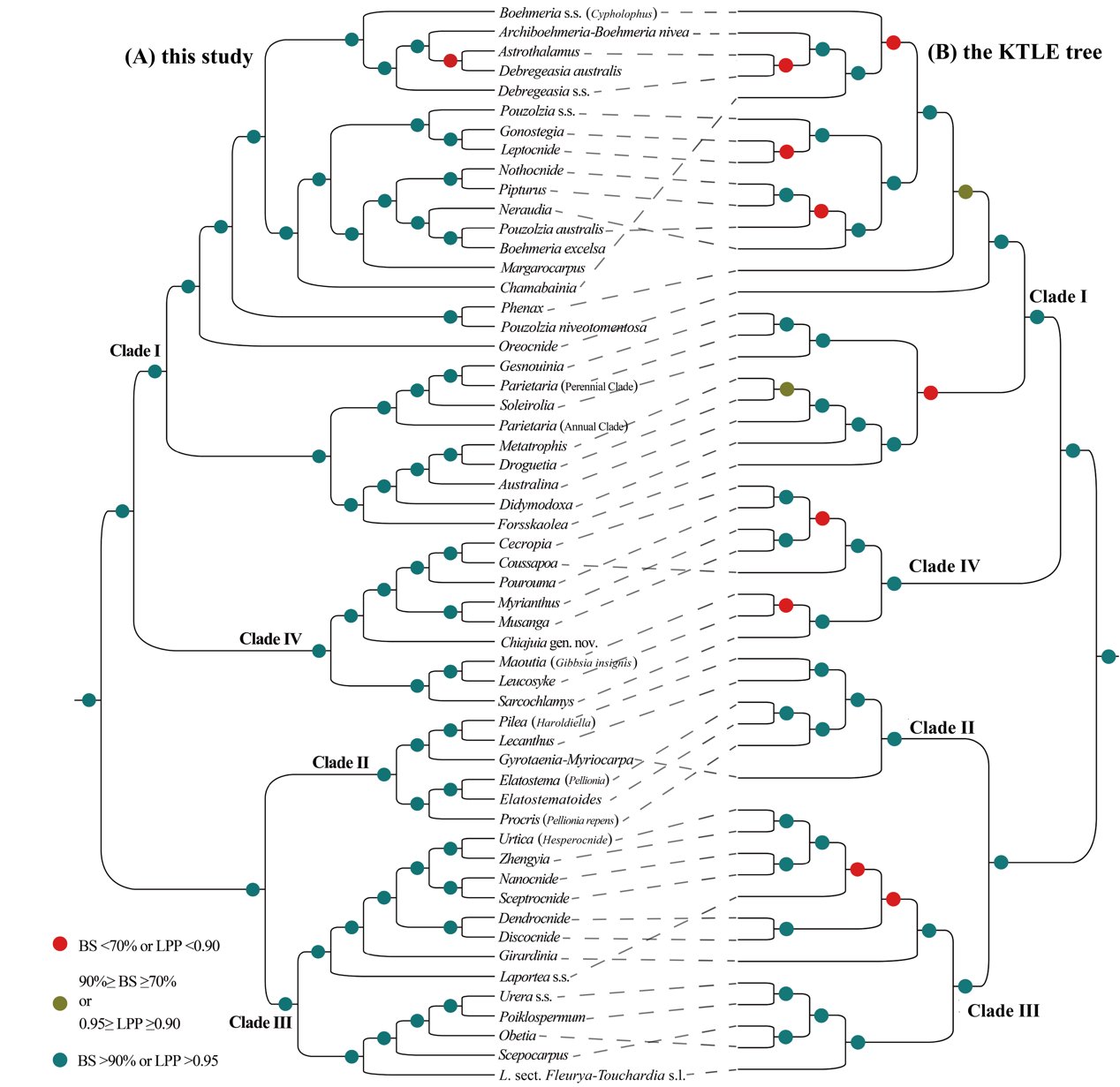
Figure 3. Comparison of phylogenetic trees constructed based on concatenated data from 87 chloroplast protein-coding genes (A) and the angiosperm "353 nuclear genes" dataset from Baker et al. (2022) (B). Black dashed lines connect identical genera or clades in both topologies. (Image by KIB)

Figure 4. Habitat and morphological characteristics of different organs of Chiajuia wallichiana (Wedd.) ZengY. Wu, X.G. Fu & D.Z. Li. A, Habitat; B, Plant habit; C, Spreading branch; D, Leaf scar and longitudinal section of stem (bottom right); E–F, Adaxial leaf surface; G, Stipule; H–I, Abaxial leaf surface; J, Arrangement of leaves and inflorescences; K, Inflorescence; L, Stamen and pedicel; M, Male flower (top left) and female flower (bottom right). (Image by KIB)
Contact:
YANG Mei
General Office
Kunming Institute of Botany, CAS
email: yangmei@mail.kib.ac.cn
(Editor: YANG Mei)




